Chapter 45 Molecular Imaging of Gene Expression and Cell Therapy
INTRODUCTION
In 1953, Watson and Crick elucidated the structure of deoxyribonucleic acid (DNA) in a 2-page letter published in the journal Nature.1 Over the next 50 years, advances in genomic science and molecular medicine have resulted in a better understanding of the pathophysiology of many diseases. Recently, another revolution has been taking place. A new set of technologies, as part of a new field termed molecular imaging, is now being developed to noninvasively examine the integrative functions of molecules, cells, and organs in intact whole-body systems.2,3 The field remains in its infancy at present and to a large extent is limited to the laboratory environment. Notable exceptions are in the disciplines of oncology, neurology, and cardiology, which have started to make the transition from basic science to clinical application. Cardiac imaging modalities such as computed tomography (CT), magnetic resonance imaging (MRI), single-photon emission computed tomography (SPECT), positron emission tomography (PET), and ultrasound have seen major advances over the past decades. In the clinical setting, these modalities can provide outstanding data regarding organ structure and physiologic function. This chapter serves as an introduction to the field of cardiovascular molecular imaging, with specific emphasis on its utility in evaluating gene and cellular therapy. Because the field may be new to many, we have purposely simplified much of the discussion on technical details to make the context more appealing to a broad range of readers with different backgrounds.
FUNDAMENTALS OF GENE EXPRESSION
The central dogma of molecular biology states that genetic information flows from DNA to ribonucleic acid (RNA) to protein. The human genome has 23 pairs of chromosomes containing approximately 3 billion base pairs of DNA and ~23,000 genes.4,5 Each gene consists of sequences of four different nucleotides: adenine (A), guanine (G), cytosine (C), and thymine (T). The flow of information from DNA to RNA within the cell nucleus is termed transcription. Each messenger RNA (mRNA) is composed of intron and exon sequences. Introns are intervening sequences that are spliced out and removed. Exons are sequences that exit the nucleus to the cytoplasm to serve as templates for protein synthesis in a process called translation. The proteins are formed from a combination of 20 different units called amino acids. Proteins are considered the workhorse of cell machinery, performing activities ranging from gene regulation to cell integrity to receptor signaling. The versatility of molecular biology cloning techniques allows both the DNA and RNA to be manipulated. For example, restriction enzymes can be used to cleave the DNA of interest at specific sites to generate discrete gene fragments.6 They are named after the bacteria from which they were isolated (e.g., Eco RI from Escherichia coli and Hind III from Haemophilus influenza) and serve originally as part of the bacterial immune arsenal to protect itself against invasion by cleaving the DNA of foreign vectors. These techniques have been used to “cut and paste” therapeutic genes and reporter genes, as will be discussed later. For an in-depth discussion on the basics of molecular biology and genomics, please refer to a recent review article.
BACKGROUND OF CARDIOVASCULAR MOLECULAR IMAGING
Traditionally, researchers have monitored cardiac gene transfer by using reporter constructs such as β-galactosidase (β-gal),7 green fluorescent protein (GFP),8 and chloramphenicol-acetyl transferase (CAT).9 Cellular transplant therapies have employed similar techniques as well as newer approaches such as TaqMan reverse transcriptase polymerase chain reaction (RT-PCR),10 Y-chromosome paint probes,11 and antibodies specific to various stem cell types.12 All of these established traditional techniques, however, require invasive biopsies and/or postmortem tissue sampling for analysis. Molecular imaging offers distinct advantages by allowing for noninvasive, quantitative, and repetitive imaging of targeted macromolecules and biological processes in living organisms.2 Although a vast array of molecular imaging techniques are available, they all require two fundamental elements: (1) a molecular probe that can signal confirmation of gene expression by detecting messenger ribonucleic acid (mRNA) transcripts or proteins and (2) a method to monitor these probes or events. Presently, the two most widely used strategies are direct and indirect imaging.
Direct molecular imaging involves direct probe-target interaction. Targets can include receptors, enzymes, or mRNA. For probe-receptor imaging, radiolabeled monoclonal antibodies binding to tumor cell-specific surface antigens have been used for the past 2 decades.13 Recent examples of cardiac application involved imaging αvβ3 integrin receptor14 or vascular endothelial growth factor (VEGF) receptor15 expressed during angiogenesis after myocardial infarction (MI). For probe-enzyme imaging, the most well-known cardiac application is fluorine-18 (18F)-labeled-fluorodeoxyglucose (FDG), used to assess for myocardial tissue viability. After transport across an intact cell membrane, the 18F-labeled glucose analog undergoes phosphorylation by hexokinase and is retained intracellularly in proportion to the rate of cellular glycolysis.16 The radioactive 18F undergoes positron annihilation into two high-energy γ rays (511 keV), which can be detected as coincidence events by PET. For probe-mRNA imaging, radiolabeled antisense oligonucleotide (RASON) probes can be used.17,18 RASON probes are typically 12 to 35 nucleotides long and are complementary to a small segment of the target mRNA. However, the RASON approach is limited at this point due to (1) low number of target mRNA (~1000 copies) per cell compared to proteins (>10,000 copies); (2) limited tracer penetration across the cell membrane; (3) poor intracellular stability; (4) slow washout of unbound oligonucleotide probes; and (5) low target-to-background ratios. On the other hand, direct imaging of DNA (two copies) within the nuclear membrane has proven exceedingly difficult and is not yet feasible. In addition, knowing the activity of gene expression (as reflected in mRNA transcripts or protein levels) rather than the number of DNA copies is more relevant for biological research. The main disadvantage of direct imaging is that it requires synthesizing a customized probe for the product (e.g., receptor, enzyme, or mRNA) of every therapeutic gene of interest, which can be time-consuming and is not generalizable to most applications.
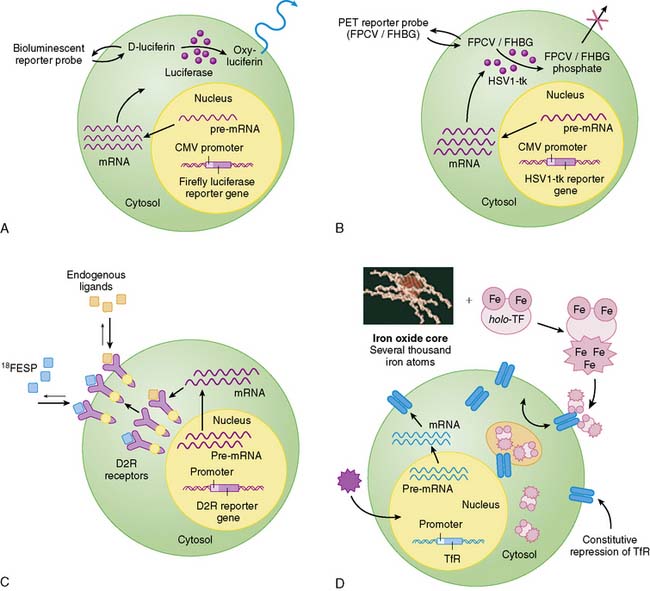
Indirect molecular imaging using reporter genes has only been recently validated. The concept of imaging reporter gene expression is illustrated in Figure 45-1. A reporter gene is first introduced into target tissues by viral or nonviral vectors. Using molecular biology techniques, the promoter or regulatory regions of genes can be cloned into different vectors to drive reporter gene mRNA transcription. Promoter activity can be “constitutive” (always on), “inducible” (turned on or off), or “tissue specific” (expressed only in the heart, liver, or other organs). Translation of mRNA leads to a reporter protein that can interact with the reporter probe. This interaction may be enzyme based or receptor-based. Probe signals can then be detected by various imaging modalities such as an optical charged coupled device (CCD) camera, PET, SPECT, or MRI.
Clearly, the main advantage of the reporter gene system is its flexibility and multiplexing capacity.19 By altering various components, the reporter gene can provide information about the regulation of DNA by upstream promoters, intracellular protein trafficking, and the efficiency of vector transduction on cells. Likewise, the reporter probe itself does not have to be changed in order to study a new biological process, saving time and resources required for synthesis, testing, and validation of new reagents. However, the main disadvantage of indirect imaging is that it remains a surrogate marker for the physiologic/biochemical process of interest, as opposed to directly measuring receptor density, mRNA copies, or intracellular enzymatic activity, which might be more clinically relevant. It should be noted that in the case of monitoring stem cell fate, this is less of a concern, since the focus is on detecting the presence of a population of cells. Finally, the ideal reporter gene and/or reporter probe should have the following characteristics:
IMAGING TECHNIQUES
Optical Imaging
Bioluminescence imaging utilizes the photogenic properties of various luciferase genes cloned from different organisms, such as firefly (Photinus pyralis), jellyfish (Aequorea), coral (Renilla), and dinoflagellates (Gonyaulax). In the case of the firefly, the firefly luciferase enzyme (FL) converts its substrate (D-luciferin) to oxyluciferin via an ATP-dependent pathway. This process emits low levels of photons (2 to 3 eV) that can be detected and counted by a cooled CCD camera (e.g., Xenogen IVIS system).20 Unlike bioluminescence, fluorescence imaging does not require injection of a reporter substrate but relies upon an excitation wavelength that produces an emission wavelength for measurement.21 Recent technologic advances (e.g., eXplore Optix system) has allowed the measurement, quantification, and visualization of fluorescence intensity in small living animals using the time domain approach.22 In general, optical-based imaging is a relatively low-cost endeavor (i.e., typically $100 to $200K versus $500K to $1 million for small-animal PET and MRI systems) with the capacity for high throughput (i.e., several mice can be scanned at once). However, the aforementioned techniques suffer from low spatial resolution, inability to monitor multiple physiologic processes, and photon attenuation/scatter within deep tissues.23 Moreover, optical imaging has yet to be extrapolated into clinical usage. Use of novel intravascular catheter devices capable of detecting bioluminescence and/or fluorescence signals from deeper tissues or organs may be possible, but the general practicality of “invasive imaging” remains to be seen.24
Magnetic Resonance Imaging
Unlike optical imaging, MRI has the advantage of a very high spatial resolution (25 to 100 μm) and the ability to measure more than one physiologic parameter at once by using different radiofrequency pulse sequences.25 This makes MR a very attractive option for imaging reporter gene expression. The imaging signal is generated as a result of spin relaxation effects, which can be altered by atoms with high magnetic moments (e.g., gadolinium and iron). One particularly useful MR imaging signal amplification system is based on the cellular internalization of superparamagnetic probes such as monocrystalline iron oxide nanoparticles (MION) and crosslinked iron oxide (CLIO).26 MIONs or CLIOs can be linked to a variety of biomolecules to produce injectable probes for targets such as hematopoietic and neural progenitor cells,27,28 activated thrombotic factor XIII,29 and endothelial cell surface markers such as E-selectin.30 These studies hold promise for in vivo imaging in humans, given the widespread availability of clinical MR scanners, nontoxic and biodegradable properties of intravenous superparamagnetic particles, and the precedent of similar preparations already in clinical use.31 However, persisting residual signals from superparamagnetic particles may hinder the capacity for quantitative and repetitive imaging. MR is also several log of orders less sensitive (10−3 to 10−5 Molar) for detection of reporter probes compared to optical bioluminescence imaging (10−15 to 10−17) or PET (10−11 to 10−12) imaging.2 Therefore, further strategies for robust signal amplification will be necessary before this modality can be of practical use for imaging cardiac gene expression and detecting small numbers of transplanted cells.
Ultrasound
The scope of ultrasound for cardiac molecular imaging has seen increasing applications over the past decade.32–34 Targeted contrast agents have been constructed by linking ligands of interest to liposomes, perfluorocarbon nanoparticles, and encapsulated microbubbles,35 but these agents are relatively large, precluding their extravasation from the vasculature. While ultrasound-based molecular imaging may play an increasing role in endothelial imaging,36 its utility as a molecular cardiac imaging modality to track gene expression or cell therapy remains to be seen. Newer techniques in photoacoustic molecular imaging where the molecular imaging agents can extravasate from blood vessels may hold a potential solution to limitations of conventional ultrasound.37
Radionuclide Imaging (See Chapter 11)
Radionuclide probes are the first example of molecular probes used in the clinical setting, and this technology represents the evolutionary roots of molecular imaging as it is known today. PET, SPECT, and planar scintigraphy have all been used to detect radionuclide-labeled probes. However, PET exhibits several advantages compared to other modalities. First, PET is more sensitive compared to SPECT and MRI for detection of probe activity, as already discussed. This may allow monitoring of gene delivery by vectors with relatively low transfection efficiencies (e.g., plasmid) or weak promoters (e.g., tissue specific), as well as detection of low numbers of cells (e.g., cardiac stem cell transplants). Second, PET imaging is more quantitative (unlike MRI) and allows for dynamic imaging with tracer kinetic modeling for analysis of rate constants underlying the biochemical processes.38 Third, inasmuch as many PET tracers have a short half-life (e.g., ~110 minutes for 18F), daily repetitive imaging of tracer retention by targeted tissues is possible. Fourth, PET imaging is tomographic (unlike two-dimensional (2D) images from optical imaging), so a relatively precise location of probe signal can be identified within the heart. This is especially apt in the basic research environment, because current generations of small-animal PET scanners have a resolution of 13 to 23 mm3 compared to around 63 mm3 for clinical PET scanners.39 Finally, studies performed in these small-animal PET scanners can potentially be scaled up to human patients using clinical PET scanners with relative ease.3
IMAGING CARDIAC GENE THERAPY
Gene transfer has been heralded as the most promising therapy of molecular medicine in the 21st century. It is usually defined as the transfer and expression of DNA to somatic cells of an individual, with a resulting therapeutic effect (see Fundamentals of Gene Expression section). In cardiovascular diseases, gene therapy offers an exciting new approach to express the therapeutic factors locally in the myocardium.40 In general, the successful application of gene therapy requires three essential elements: (1) a vector for gene delivery, (2) delivery of the vector to the target tissue, and (3) a therapeutic gene to be expressed in a particular patient population.
These encouraging results have led to the initiation of several clinical trials, beginning in the 1990s. Of the 509 ongoing gene therapy trials in the United States, 46 are related to cardiovascular diseases. The majority of these are aimed at testing the safety and efficacy of therapeutic angiogenesis and, to a lesser extent, examining restenosis. In the late 1990s, several phase-1 open-labeled trials involving small numbers of patients with myocardial ischemia and peripheral vascular disease uniformly showed positive results.48–50 However, recent phase-2 randomized, double-blind, placebo-controlled trials have yielded conflicting, if not disappointing, results. The Vascular Endothelial Growth Factor in Ischemia for Vascular Angiogenesis (VIVA),51 FGF Initiating Revascularization Trial (FIRST),52 Adenovirus Fibroblast Growth Factor Angiogenic Gene Therapy (AGENT),53,54 and Kuopio Angiogenesis Trial (KAT)55 tested gene therapy using either VEGF or FGF. Unfortunately, these trials failed to show any consistent improvement in various parameters such as symptomatic improvement, ejection fraction, wall motion scores, myocardial perfusion, and restenosis rate.
Nonetheless, important lessons can be learned from these trials. They showed that angiogenesis is a complex process regulated by the interaction of various growth factors and may be difficult to stimulate using a single protein or gene injection. The ideal injection method, delivery vector, and patient population remain to be determined. The pharmacokinetics and pharmacodynamics of therapeutic gene expression will need to be defined first before gene therapy can proceed further to widespread clinical usage, similar to the research and development of experimental drugs.56 Finally, since there is no available method of assessing gene expression in vivo, the investigators are unable to determine whether the lack of symptomatic improvement is due to poor injection technique, insufficient gene expression, host inflammatory response, or an inappropriate therapeutic gene.
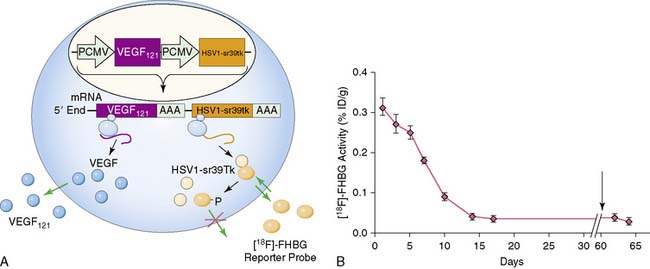
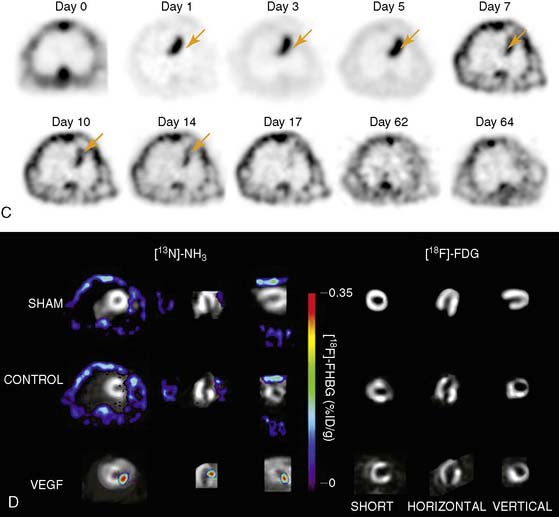
Imaging of cardiac transgene expression has been established in several proof-of-principle studies involving injection of various reporter genes into the myocardium and following the kinetics of transgene expression over time, using optical bioluminescence, microPET, and clinical PET imaging.57–61 The feasibility of linking a PET reporter gene to a therapeutic gene was also demonstrated in two separate studies.62,63 In this case, an adenovirus containing two constitutive cytomegalovirus promoters driving a VEGF121 therapeutic gene and an HSV1-sr39tk PET reporter gene separated by polyadenine sequences was constructed (Ad-CMV-VEGF121-CMV-HSV1-sr39tk). Wu et al. injected the construct into the myocardium of a rat in a myocardial infarction model.63 Reporter gene expression (which indirectly reflects the VEGF121 therapeutic gene expression) persisted for only about 2 weeks, owing to host immune response against the adenovirus. At 2 months, there were no significant improvements in myocardial contractility, perfusion, or metabolism, as measured by echocardiography, nitrogen-13-labeled ammonia (13N-NH3), and FDG imaging, between study and control groups (Fig. 45-2). Thus, this study highlights the importance of monitoring the pharmacokinetics of gene expression. It also demonstrates the proof of principle that any other cardiac therapeutic genes of interest (e.g., HIF-1α, SERCA2a, heat shock protein, or endothelial nitric oxide synthase) can likewise be coupled to a PET reporter gene for subsequent noninvasive monitoring.