Lung Immunology |
The lung is constantly exposed to foreign agents. As a result it is faced with the challenge of distinguishing nonpathogenic moieties in ambient air from potentially pathogenic antigens derived from microorganisms. Here, we use the term “pathogens” to refer to infectious agents, allergens, toxins, and other inhaled antigens unless stated otherwise. The lung protects itself using local tissue structures such as the mucus layer, ciliary ladder, and smooth muscles. It also employs complex immune defenses that are both innate and adaptive.1,2 The immune responses need to be able to recognize and to react to a wide variety of stimuli. They must be able to recognize and to eliminate unwanted pathogens to keep pulmonary structures free of infection. On the other hand, they must not overreact to inhaled stimuli to avoid potential excessive inflammation and lung injury. This need to control the intensity and duration of such responses is required to preserve normal lung structure, especially the highly vascularized and fragile alveolar epithelial surface that is required for gas exchange. Alterations of these lung protective mechanisms lead to many of the pulmonary diseases physicians face in their patients. Therefore, understanding the innate and adaptive immune responses in the lung is important in our attempts to understand the pathophysiology and to improve the management of many pulmonary diseases. The anatomic and immune defenses will be reviewed in this chapter.
ANATOMIC MECHANISMS
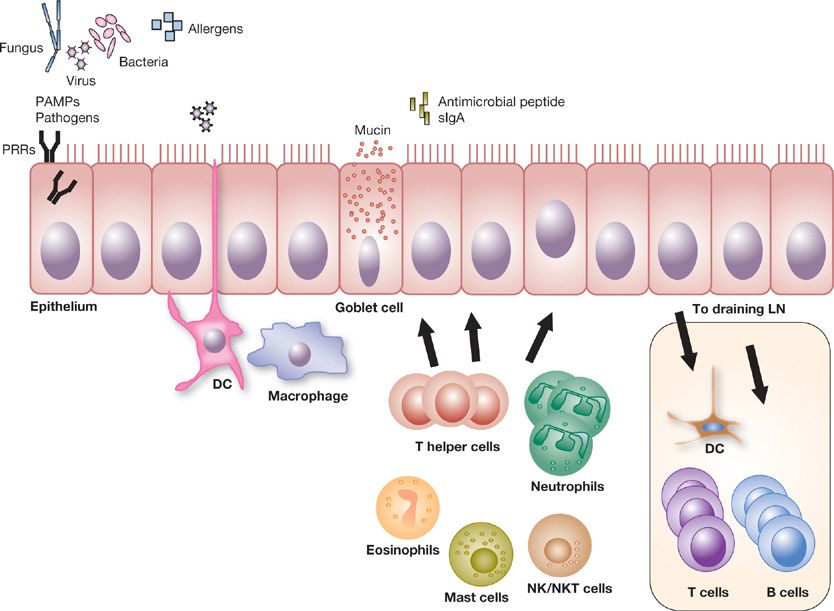
Given the lungs’ large surface area, it is exposed to many inhaled environmental challenges, because the air we breathe contains infectious agents, toxic gases, and fine particulate matter (Fig. 20-1). The alveolar and capillary membrane barriers are important for gas exchange, and need to be defended from the injurious effects of incoming toxic and infectious pathogens. If the consequences of these exposures are not controlled, this can lead to excessive inflammation, lung edema, as well as propagation of infectious agents. This can, in turn, lead to alveolar destruction, abnormal fibrotic repair, and compromised gas exchange. Air is inhaled through the nose or the mouth into the extrathoracic portion of the trachea before it enters the thorax. The nose filters and conditions the inhaled air for humidity and body temperature as it flows through the nasal turbinates. Nasal hairs also provide a barrier that traps larger particulates. The nasal secretions lining the airway mucosa contain many substances such as lysozyme, immunoglobulins (such as secretory immunoglobulin A [sIgA]), and antimicrobial peptides that bind to, and inactivate invading microbes. For example, sIgA accounts for 15% of the total protein in upper airway secretions and plays a significant role in neutralizing and preventing epithelial attachment of invading viruses and bacteria.3 The conducting airway mucosa is also coated with acidic viscous fluid and mucus secreted by Clara cells, goblet cells, and bronchial glands (Fig. 20-1). This fluid makes up an important airway surface lining that coats the bronchial epithelium and forms a barrier between the outside world and the lung parenchyma.
Figure 20-1 Innate recognition of pathogen or PAMPs on airway epithelium. Pathogens such as fungus, virus, or bacteria (or their associated pattern-associated molecular proteins [PAMPs]) and environmental allergens are first detected by membrane bound, cytosolic and/or endosomal innate sensors or pattern recognition receptors (PRRs). The respiratory epithelial cells provide a physical barrier between the airway luminal antigens and the underlying respiratory tissues in their control of immune defense and tolerance. Specialized epithelial cells can have cilia, produce a mucus layer and secrete antimicrobial proteins and secretory IgA (sIgA) that limit pathogen exposure to the epithelial cells. Dendritic cells (DCs) interdigitating between and lying near the epithelium, as well as tissue macrophages, are well positioned to sample and ingest incoming antigen. Recognition of the antigen by the innate immune receptors leads to production of pro-inflammatory cytokines and chemokines which recruits immune cells such as neutrophils, T cells, NK cells, NKT cells, eosinophils, or mast cells. Activated DCs that have taken up the inhaled antigen migrate to nearby draining lymph nodes. In the lymph nodes, the DCs help integrate the signals from the inhaled substance and the innate immune response into signals that direct cells of the adaptive immune system. The DCs encounter appropriate naïve CD4+ T cells where factors such as the phenotype of the antigen-presenting cells (APCs) and cytokine milieu modulate differentiation of CD4+ T cell subsets with their characteristic cytokine and functional profiles. These immune cell functions and cytokine production determine whether an activating or a tolerant response will develop.
INNATE IMMUNITY IN THE LUNG
The innate immune response is an evolutionarily conserved system of host defense evident in all multicellular organisms. In the constant battle between host and pathogens, the purpose of the innate immune system is to provide a rapid response, in contrast to the adaptive immune system, which provides a more specific response but takes longer to respond (see Section “Adaptive Immune Responses in the Lung”). Because humans breathe approximately 10 L of air per day (12 to 15 breaths/min × 500 mL tidal volume), we are constantly exposed to pathogens (e.g., viruses, bacteria, and fungi), allergens (e.g., house dust mites, dander), and toxins (e.g., cigarette smoke and pollutants). The innate immune system allows humans to respond to these stimuli even if they have not been exposed to them previously.
Because of the constant exposure to the environment, similar to other mucosal surfaces (e.g., gut and skin), the innate immune system must maintain a balanced response. Ideally, this should include (1) pathogen recognition, (2) initiation of an appropriate response, which is typically manifested by inflammation, and (3) resolution of inflammation. An inappropriately regulated innate response contributes to the pathogenesis of lung diseases as diverse as asthma, emphysema, and interstitial lung disease.
The most important property of the innate immune system is its response to pathogens regardless if there has been a prior exposure. Thus, an acquired immunologic memory against the pathogen is not required. The anatomic and antimicrobial structures that prevent infection and injury noted earlier can be thought of as part of the innate response. Therefore, one of the most important roles of the lung innate immune system is its function as a barrier to inhaled pathogens.4 The epithelial surface of the lung creates a barrier with tight junctions between neighboring cells that prevent pathogen entry (Fig. 20-1). In addition to this barrier function, the epithelium has a coordinated system for the removal of inhaled pathogens. Goblet cells and secretory glands produce mucus that engulfs pathogens. A coordinated system of ciliated cells moves mucus up the mucociliary “ladder” to be expectorated during coughing. A disturbance in this essential function leads to significant pathology. In primary ciliary dyskinesia (PCD), cilia function is impaired and individuals develop bronchiectasis with recurrent bacterial infection.5 In cystic fibrosis (CF), an abnormality in epithelial chloride channel function causes changes in the airway surface liquid that prevents effective mucus clearance, which also leads to chronic inflammation, bronchiectasis, and recurrent bacterial infections.6,7 The mucociliary “ladder” is particularly effective for large molecules, but smaller molecules (<5 μM) are able to directly descend to the distal alveolar epithelium. Here, instead of mucociliary clearance, particles encounter secreted surfactant proteins (specifically SP-A and SP-D; also known as pulmonary collectins) that act as opsonins, and assist resident macrophage in phagocytosis.8 Finally, the epithelium also produces a variety of innate immune antimicrobial molecules that activate alveolar macrophages (AMs) to kill pathogens.9
 PATHOGEN RECOGNITION
PATHOGEN RECOGNITION
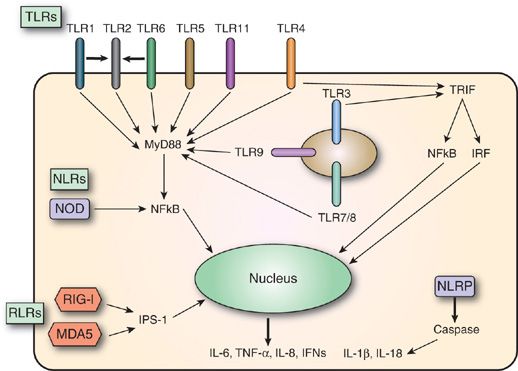
The innate immune response allows the host to respond to a wide array of pathogens. However, the mechanism(s) that the host uses to recognize these pathogens was poorly understood until the 1980s. At that time, a seminal advance in our understanding of the immune system occurred when it was proposed that the host detects, and responds to these pathogens via a cadre of receptors that recognize conserved sequences called pathogen-associated molecular patterns (PAMPs), which exist in pathogens, and are not present in the host (Fig. 20-2).10,11 Experiments in Drosophila (fruit flies) in 1996 identified pattern recognition receptors (PRRs) that recognized PAMPs and supported the concept of innate receptors.12 Subsequently, PRRs were identified in humans,13 and more recently, additional PRRs have been shown to recognize endogenous molecules released from damaged cells, which are referred to as damage-associated molecular patterns (DAMPs).14,15 The major types of PRRs are described in sections “Toll-Like Receptors”, “RIG-I-Like Receptors”, “Nod-Like Receptors” and “C-Type Lectin Receptors”.
Figure 20-2 Pattern recognition receptors (PRRs). Classes of PRRs include Toll-like receptors (TLRs), retinoic acid–inducible gene-I–like receptors (RLRs) and nucleotide oligomerization domain–like receptors (NLRs). Not shown are C-type lectin receptors (CLRs). TLRs, RLRs (e.g., RIG-I, MDA5, and other sensors), and NLRs (e.g., NOD1, NOD2, NLRP3, etc.) are innate immune sensors that recognize danger signals derived from pathogens (PAMPs), damaged cells (DAMPs), or associated nucleic acids at the cell surface, in endolysosomes or in the cytoplasm. Signaling by these sensors promotes, either the activation and nuclear translocation of transcription factors (IRFs, NFκB and AP-1) that drive expression of cytokines (IFN-α/β, TNF, and pro-IL-1β), or the assembly of the caspase-1 inflammasome and subsequent maturation of IL-1β from pro-IL-1β. IFN, interferon; IL, interleukin; IRFs, interferon regulatory factors; MDA5, melanoma differentiation–associated gene-5; NFκB, nuclear factor κB; NLRP, NLR with a pyrin domain; NOD, nucleotide oligomerization domain; RIG-I, retinoic acid–inducible gene-I; TNF, tumor necrosis factor; TRIF, TIR-domain–containing adapter-inducing interferon-β.
Toll-Like Receptors
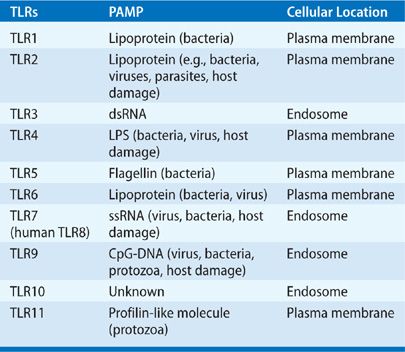
The mammalian Toll-like receptor (TLR) family received its name from the Drosophila Toll gene. Early studies that involved the genetic manipulation of Toll resulted in dramatic defects in innate immune responses (Table 20-1; Fig. 20-2).12,16 Since this discovery, the TLRs are appreciated to be the prototypical PRR molecules.13,17 It is now known that some TLRs are present on the cell surface, and others are in endosomes (intracellular vesicles). As a group, they are characterized by N-terminal leucine-rich repeats that recognize pathogens, a transmembrane region, and a cytoplasmic domain that is highly homologous to the interleukin (IL)-1 receptor and the IL-18 receptor.18 To date, 11 TLRs have been identified in humans, with different TLRs recognizing distinct PAMPs.19 In general terms, TLR recognition of PAMPs is based upon cellular compartmentalization. Recognition of viral RNA and DNA occurs in endosomes by TLR3, TLR7, and TLR9, while TLR2 and TLR4 have been shown to recognize viral proteins. The remaining TLRs recognize bacteria, parasite, and host proteins (e.g., DAMPs), while TLR7 and TLR9 recognize DNA from bacteria or protozoa.
TABLE 20-1 Toll-Like Receptors
TLR Signaling Pathways
TLR recognition of PAMPs stimulates signaling pathways that ultimately lead to activation of gene transcription and protein production. The responses that are elicited can be cell type specific. They also utilize two distinct pathways based on which of two adaptor molecules they employ: (1) myeloid differentiation primary response gene 88 (MyD88) and (2) TIR-domain–containing adapter-inducing interferon-β (TRIF). With the exception of TLR3, and one of the pathways downstream from TLR4, MyD88 is required for the downstream signaling of TLRs. MyD88 interacts with the IL-1R–associated kinase (IRAK)-4, and other IRAK family members, before they dissociate from MyD88 and interact with TNFR-associated factor 6 (TRAF6). IκB kinase (IKK)-β and MAP kinases are activated, leading to another complex of IKK molecules that are degraded and release nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB). NF-κB is a critical transcription factor for TLR-induced genes in inflammation. In addition, activation of the MAP kinase cascade leads to another transcription factor, AP-1, which is associated with activation of cytokines. TRIF-dependent signaling was found to be responsible for TLR3 signaling and, more recently, TRIF was implicated in TLR4 signaling.20 TRIF interacts with TRAF3 and TRAF6, and leads to TNF receptor (R)-associated death domain protein (TRADD), ultimately leading to activation of NF-κB. Interestingly, TLR4 was found to activate MyD88 at the surface of the plasma membrane, while TRIF activation occurred when TLR4 was in endosomes.20 Downstream from TRIF is activation of interferon regulatory factors (IRFs), which lead to interferon production and activation of interferon-dependent genes.
RIG-I–Like Receptors
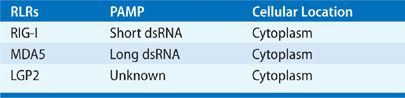
The RIG-I–like receptor (RLR) family presently includes three members: retinoic acid–inducible gene-I (RIG-I), melanoma differentiation–associated gene 5 (MDA5), and laboratory of genetics and physiology 2 (LGP2) (Table 20-2).21 RLRs are expressed at a basal level by all nucleated cells, suggesting a central role in innate immune recognition and response to viral infection. A measure of their importance can be seen in the evolution of viral mechanisms to evade or suppress RLRs.22 In the 1950s, interferons were discovered to be a critical antiviral innate immune response to viral infection produced by infected cells, and important in the recruitment and activation of natural killer (NK) cells and lymphocytes that recognize and kill virus-infected cells.23 While the presence of an intracellular TLR receptor for dsRNA was hypothesized in 1969,24 and subsequently identified, the discovery of RLRs introduced a second family of PRRs that are cytoplasmic sensors of viral nucleic acids that contribute to antiviral host defense by inducing interferon production. RIG-I was initially discovered in 1997 as a gene induced in acute promyelocytic leukemia cells after stimulation with retinoic acid.25,26 Several years later, RIG-I was found to be activated by interferon.27 Similarly, MDA5 was identified in 1999,28 and was later found to be induced by interferon.29 LGP2 was suggested to function as a RIG-I inhibitor, exerting a regulatory role on antiviral innate immune responses,30 but recent experiments suggest that LGP2 may modify viral RNA to assist RIG-I and MDA5 recognition of dsRNA.19 Three genes encode RLRs in mice and humans, and all share a conserved helicase domain. Briefly, RIG-I and MDA5 are composed of two N-terminal caspase recruitment domains (CARDs), a central DEAD box helicase/ATPase domain, and a C-terminal regulatory domain.31 LGP2 retains the helicase domain but lacks the CARD domain.31 Their cytoplasmic location allows for recognition of dsRNA viruses and dsRNA intermediates generated during ssRNA viral replication. In addition, there is evidence that ssRNA is detected from viruses that do not produce significant amounts of dsRNA.19 Importantly, eukaryotic cells avoid self-activation of RLRs by RNA in mitochondria, and RNA released into the cytosol is packaged to avoid RLR recognition.22
TABLE 20-2 RIG-I-Like Receptors
In the human lung, RLRs have been found to respond to a range of respiratory viruses, from rhinovirus (common cold virus) to influenza virus. In response to viruses, activation of intracellular signaling ultimately leads to interferon production and the activation of interferon-dependent genes. RLR signaling is dependent on CARDs, which interact with the interferon-β–promoter stimulator 1 (IPS-1, also known as MAVS, CARDIF, or VISA). IPS-1 is localized on the mitochondrial membrane. IPS-1 activates TRAF3 and TRADD, which are common molecules for IFN-inducible gene expression.19 In addition, NF-κB signaling has been implicated.32
NOD-Like Receptors
Nucleotide oligomerization domain (NOD)-like receptors (NLRs) are another family of cytoplasmic pathogen sensors that were discovered in the 1990s (Table 20-3).33 Briefly, the molecular structure of NLRs includes a central nucleotide-binding domain and C-terminal leucine-rich repeats. The N-terminal portions of most NLRs contain protein-binding motifs (e.g., CARDs, a pyrin domain, and a baculovirus inhibitor of apoptosis protein repeat [BIR] domain).19 NLRs activate important signaling pathways mediated by NF-κB and MAP kinases, and activate caspases. The result is activation of diverse signaling pathways that induce innate immune responses.34

NOD1 and NOD2 identify bacterial PAMPs by recognition of peptidoglycan from gram-negative bacteria, gram-positive bacteria, and mycobacteria. The importance of NLRs was highlighted by the evidence that NOD polymorphisms are associated with increased severity of atopy, eczema, and asthma,35 as well as other inflammatory diseases (e.g., Crohn disease,36 tuberculosis,37 and lung cancer38). This suggests that a defect in bacterial clearance may influence the host microbiome and subsequent pathophysiology of disease. Distinct from NOD1 and NOD2 are a group of NLRs that activate caspases, such as caspase-1. Caspase-1 is the prototypical caspase that has been shown to be required for cleavage and processing of inflammatory cytokines (IL-1β, IL-18) into their active forms. This signaling pathway is now commonly referred to as the “inflammasome,” and has been shown to be critical for effective innate immune responses against a wide range of microbial pathogens, cancer, and inflammatory, metabolic, and autoimmune diseases.39 Specific to the lung, the inflammasome is critical for responses to important pathogens such as influenza A virus,40,41 Streptococcus,42,43 Pseudomonas aeruginosa,44 and Mycobacterium tuberculosis.45 In addition, activation of the inflammasome has been suggested to contribute to the pathogenesis of asthma,46 chronic obstructive pulmonary disease (COPD),47 and pulmonary fibrosis.48
C-Type Lectin Receptors
The C-type lectin receptors (CLRs) were identified in the early 1900s, and represent a diverse family that includes over one thousand members.49 The CLRs were initially characterized as Ca2+-dependent (C-type) carbohydrate-binding (lectin) proteins49 but, subsequently, some CLRs were found to not bind carbohydrate ligands or require Ca2+-dependent signaling.50 Currently, CLRs have been classified into 17 groups.50 Structurally, CLRs are characterized by a carbohydrate-recognition domain (CRD) that is highly conserved.49 Despite this conserved CRD structure, CLRs are involved in cell functions as diverse as adhesion, repair, endocytosis, and phagocytosis, as well as innate immune pathogen recognition. CLRs recognize carbohydrates on a wide variety of pathogens including viruses, bacteria, and fungi. Several of the canonical CLRs include dectins, mannose-binding lectin (MBL) receptors, and surfactant protein (SP). Dectin-1 and Dectin-2 are important PRRs for recognition of fungi.51 Upon recognition of fungal PAMPs, intracellular signaling pathways involve activation of NF-κB and MAP kinases.52 MBL receptors function as PRRs by recognition of carbohydrate motifs on pathogens, which leads to complement activation.18 MBL recognizes diverse pathogens that include gram-positive and gram-negative bacteria, yeast, parasites, mycobacteria, and viruses.53 SP was first identified as a critical phospholipid for maintaining alveolar compliance to prevent atelectasis.54 However, in the 1980s after the discovery of MBLs, sequencing of SP revealed significant structural similarities.55 In accord with these similarities, SP-A and SP-D, secreted by alveolar epithelium, have been shown to function as agglutinins, opsonins, and inflammatory modulators8 in response to viruses, bacteria, mycobacteria, and fungi.56
 PATHOGEN-INDUCED INNATE IMMUNE RESPONSES
PATHOGEN-INDUCED INNATE IMMUNE RESPONSES
Inhaled pathogens may circumvent the mucociliary “ladder” on airway epithelium, which is typically the first responder to pathogens by virtue of its location and distribution. Antigen-presenting cells (APCs) such as dendritic cells (DCs) provide a critical role for pathogen recognition by extending projections into the airway between epithelial cells to sample the environment. On both airway epithelium and DCs, there is a wide repertoire of PRRs that recognize pathogens.57 Upon recognition of PAMPs, PRRs activate signaling pathways that lead to subsequent inflammatory responses. An important component of this inflammatory response is the production of chemokines to recruit hematopoietic cells (e.g., neutrophils, eosinophils, macrophages, and lymphocytes including NK, and NKT cells) that are critical effector cells for the initiation and resolution of inflammation. In addition to chemokine production, PRR activation stimulates the production of a variety of cytokines (e.g., tumor necrosis factor (TNF)-α, IL-1β, IL-6, TGF-β, IL-10) that activate effector cells and contribute to the regulation of innate and adaptive responses in the lung.57 PRR activation influences the balance of oxidation and proteases present in the lung. Importantly, dysregulation of oxidation has been shown to contribute to the pathogenesis of COPD,58 while defective antiproteases, best exemplified by α1-antitrypsin deficiency, is a significant risk factor for the development of asthma, bronchiectasis, and emphysema.59 Antimicrobial peptides are also produced by epithelial cells, as well as recruited hematopoietic cells, in response to PRR activation (Fig. 20-1).60 In humans, two significant groups of antimicrobial peptides are the cathelicidins61 and defensins.60 These small cationic peptides have antimicrobial activity against gram-negative and gram-positive bacteria, fungi, parasites, and viruses.62 Epithelial and hematopoietic cells (e.g., neutrophils) produce both cathelicidins and defensins, and in addition to their antimicrobial properties, these peptides have been implicated in the induction of inflammation, influencing adaptive immune responses and wound repair.61,62 Therefore, antimicrobial peptides are an excellent example of an innate immune response, initiated by PRR recognition of pathogens or PAMPs, which results in a protective response, regulates inflammation, and bridges innate and adaptive immunity. Recently a group of innate immune cells, known as innate lymphoid cells (ILCs), have been identified that lack specific antigen receptors but can produce an array of effector cytokines.63,64 These ILCs have morphologic characteristics of lymphoid cells and can respond to a variety of signals and play an important role in immunity against microorganisms, tissue homeostasis, and repair of damaged tissues. Currently, there are three subsets of ILCs identified: ILC1s include IL-15–dependent NK cells; ILC2s are characterized by their production of T helper (Th) 2 cytokines (IL-5 and IL-13); and ILC3s express the nuclear hormone receptor retinoic acid receptor–related receptor (ROR)γt and produce cytokines IL-17 and IL-22.63,65 These cells are important in innate immunity and very likely will be discovered to contribute to the development of effective adaptive responses.
CELLULAR RESPONSES IN THE LUNG
When pathogens bypass the initial lung mucosal barriers, the host’s immune system responds in an orchestrated defense that involves a number of specialized cells that target the threat, neutralize it, and cleanup remnants to prevent the tissue injury. Macrophages function as phagocytes to engulf pathogens (viable and nonviable microorganisms), as well as apoptotic and necrotic cells that have undergone physiologic and pathologic cell death.66 Ultimately, the lung must perform these tasks in a selective manner to avoid unnecessary inflammation that can cause continued tissue destruction. The innate immune system, as described earlier, is made up of a humoral arm (lactoferrins, lysozyme, sIgA, SPs, MBLs, and defensins) as well as a cellular arm (AMs, DCs, neutrophils, ILCs, etc.) that express numerous PRRs and or phagocytic receptors important for their diverse functions.
 MACROPHAGES
MACROPHAGES
Macrophages, initially described in the 1880s, are large mononuclear phagocytic cells. They play an important role in host defense based on their ability to phagocytize inhaled moieties, maintain tissue homeostasis, and function as APCs which are important in adaptive humoral and cell-mediated responses.67 Macrophage precursors arise from committed hematopoietic stem cells in the bone marrow and are released into the circulation as monocytes before they differentiate into macrophages and DCs as they migrate into the lung. Tissue-resident macrophages form a specialized population based on their local anatomic location. Once called dust cells for their ability to engulf particulates, AMs, named for their presence in the pulmonary alveolus, frequently contain granules of various exogenous materials.68 One example is particulate matter such as the black carbonaceous granules seen in lungs from smokers and long-term city dwellers. Routine bronchoalveolar lavage will identify these resident airway and AMs. However, pulmonary interstitial macrophages from lung tissue can only be collected by tissue dispersion techniques.69 Of note, AMs are inferior APCs when compared to DCs.70
AMs secrete numerous products (e.g., cytokines, chemokines, and peptides) and directly interact with other cells and molecules through the expression of a variety of surface receptors.71 AMs have multiple functions that include phagocytosis of cells undergoing apoptosis and necrosis, clearance of ingested pathogens or particulates, routine clearance of surfactant, and suppressing inappropriate inflammation and immune responses to harmless inhaled antigens. Macrophages ingest and phagocytose microbial or environmental particulate matter, which is then enclosed in intracellular vesicular phagosomes that undergo fusion with primary or secondary lysosomes to form phagolysosomes. In these AMs vacuoles, intracellular killing takes place through mechanisms that include antimicrobial proteins, degradative enzymes, oxidation, reactive oxygen intermediate generation, and “respiratory burst” known as nicotinamide adenine dinucleotide phosphate (NADPH) oxidase degradation. Macrophages facilitate many innate recognition functions because they express diverse PRRs (e.g., TLRs, NLRs, and CLRs) to assist host defense in combating invading pathogens. The recognition and clearance of invading pathogens can also occur via opsonin-mediated phagocytosis through the Fcγ receptors. These receptors allow macrophages to be selective in their response to phagocytosed antigen, minimize damaging effects on normal cells, and limit responses to harmless moieties that can be frequently encountered in the airway.
While AMs have greater respiratory burst capacity than other phagocytes,72
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree