Fig. 11.1
(a) P. jirovecii asci visualized in a patient’s bronchoalveolar lavage using Grocott’s methenamine silver staining as described by Churukian and Schenk (1977). The parentheses-like structure in each ascus resembles opercula observed in other asci. Ascomycetous operculum is thought to be involved in the dehiscence (release of endospores) upon contact with humidity, suggesting that asci might be the airborne P. jirovecii infectious forms (picture from Institute of Microbiology, Centre Hospitalier Universitaire Vaudois). (b) P. jirovecii organisms visualized in a patient’s induced sputum using direct immunofluorescence kit BioRad MONOFLUO Pneumocystis (Picture from Critical Care Medicine Department, NIH Clinical Center, National Institutes of Health)
Table 11.1
Number of enzymes dedicated to the biosyntheses of amino acids in P. jirovecii, P. carinii, and S. pombe
Amino acid | Number of dedicated enzymes | |||
|---|---|---|---|---|
In S. pombe (reference)a | In S. pombe | In P. jirovecii | In P. carinii | |
Ala | 1 | 1 | 1 | 0 |
Asp | 1 | 1 | 1 | 1 |
Asn | 1b | 1 | 0 | 0 |
Arg | 3 | 3 | 0 | 0 |
Cys | 2 | 1 | 0 | 0 |
Glu | 1b | 1 | 1 | 1 |
Gln | 1 | 1 | 1 | 0 |
Gly | 1b | 1 | 1 | 0 |
His | 6 | 6 | 0 | 0 |
Ilec | 4 | 4 | 1 | 0 |
Leud | 3 | 3 | 1 | 0 |
Lys from aspartate | 0 | 0 | 0 | 0 |
Lys from pyruvate | 7 | 3 | 0 | 0 |
Met | 3 | 2 | 0 | 0 |
Phe | 2 | 1 | 0 | 0 |
Pro | 1b | 1 | 0 | 0 |
Ser | 3 | 3 | 1 | 0 |
Thr from glycine | 1 | 1 | 0 | 0 |
Thr from homoserine | 2 | 2 | 0 | 0 |
Trp | 5 | 3 | 0 | 0 |
Tyr | 2 | 1 | 0 | 0 |
Valc | 4 | 4 | 1 | 0 |
Total | 54 | 44 | 9 | 2 |
III. Epidemiology
Immunity against P. jirovecii develops during the first years of life, presumably after a primary infection that occurs prior to 2 years of age (Pifer et al. 1978; Vargas et al. 2001). Thus, carriage of the fungus as a latent infection and reactivation upon immunosuppression was the first hypothesis for the development of PCP later in life. However, a number of recent observations have suggested that infection de novo from exogenous sources is responsible for the majority of episodes: (i) the occurrence of PCP outbreaks caused by a single genotype (de Boer et al. 2011), (ii) the elimination of Pneumocystis after experimental infection in the rat model (Vargas et al. 1995), and (iii) the presence of mutations within the dihydropteroate synthase gene in patients who never received sulfa drugs (Huang et al. 2000; Nahimana et al. 2003a). The latter observation provides especially strong evidence supporting de novo infection because these mutations (i) are associated with failure of sulfa prophylaxis, (ii) are absent in P. jirovecii isolated before the use of sulfa drugs for treating or preventing PCP (Meshnick 1999), and (iii) have been shown to be selected by sulfonamide use within single patients (Nahimana et al. 2003b).
The occurrence of infection de novo raises questions about the source of the pathogen and mode(s) of transmission. Obligate parasitism implies that the reservoir and source of infection are probably only humans.
A number of experiments in animal models have helped in understanding the epidemiology of P. jirovecii in humans. Studies on rats demonstrated that air is the route of transmission for de novo infection (Hughes 1982; Hughes et al. 1983). Horizontal transfer of the fungus can occur from animals with PCP to susceptible animals (Powles et al. 1992; Vogel et al. 1993; Ceré et al. 1997), but also from immunocompetent murine carriers of the fungus following a single day of exposure (Dumoulin et al. 2000; Chabé et al. 2004).
Carriage of P. jirovecii without overt PCP by immunocompromised (Nevez et al. 1999; Hauser et al. 2000) and immunocompetent (Vargas et al. 2000; Miller et al. 2001; Tipirneni et al. 2009) humans has been documented. The genotypes observed in carriers (Hauser et al. 2000) and in infants, presumably experiencing primary infection (Totet et al. 2003, 2004), were not different from those in PCP patients, suggesting that these could be sources of the pathogen. However, one study of larger cohorts found that infants may harbor different genotypes from AIDS patients (Beard et al. 2005). This study suffers from the fact that the infant and adult populations were from different geographical areas, which may have had different predominant genotypes. The dispersion of Pneumocystis organisms in the air around PCP patients (Choukri et al. 2010; Bartlett et al. 1997), as well as around infected rats (Choukri et al. 2011) has been examined by PCR techniques. Significant fungal burdens were detected in the air at 1 m from the source, then decreased with distance but were detected up to 8 m away. For rats, air burdens correlated with lung burdens. The same P. jirovecii genotypes were present in the lungs of PCP patients and in air particles collected in their room (Bartlett et al. 1997; Damiani et al. 2012). Together, these observations suggest that inter-human transmission from infected humans through the air is the main route of dissemination of P. jirovecii. The possible categories of humans that may act as a source include patients with active or developing PCP, infants experiencing primary infection, and colonized immunocompromised patients, but also, possibly, transient carriers such as elderly people (Vargas et al. 2010), pregnant women (Vargas et al. 2003), and health-care workers (Miller et al. 2001; Tipirneni et al. 2009). Nevertheless, the actual human source(s) of infection for sporadic cases and their relative importance remain to be elucidated. We speculate that infants experiencing primary infection might play a major role because, being immunologically naive, multiplication of the fungus might be more productive than in other categories of colonized humans.
IV. Molecular Methods for Investigating PCP Outbreaks
In order to test the hypothesis that inter-human transmission of P. jirovecii is involved in outbreaks, molecular typing methods are crucial because they can document identical P. jirovecii genotypes in patients between whom transmission is suspected. Typing methods for P. jirovecii rely on the natural genetic heterogeneity among isolates. In the absence of a culture method, current typing methods utilize PCR amplification of variable regions of the P. jirovecii genome, followed by the detection of polymorphisms by DNA sequencing, single-strand conformation polymorphism (SSCP) analysis, or restriction fragment length polymorphism (RFLP) analysis. Initially, one to two genetic regions were sequenced, including a variable region of the mitochondrial 26S rRNA gene (hereafter called mt26S) (Keely et al. 1995) or the internal transcribed spacers number 1 and 2 of the nuclear rRNA operon (ITS1 and ITS2; ITSs sequencing) (Lee et al. 1993; Lu et al. 1994; Tsolaki et al. 1996). Subsequently, a multilocus approach was developed that utilized SSCP to analyze four genomic regions (Hauser et al. 1997, 2001a; Nahimana et al. 2000), including ITS1, mt26S, the intron of the nuclear 26S rRNA gene (26S), and the β-tubulin intron 6 region (b-tub). As DNA sequencing prices decreased, SSCP was replaced by direct sequencing, i.e., without cloning PCR products (multilocus sequence typing, MLST) (Schmoldt et al. 2008; Gianella et al. 2010; Pliquett et al. 2012). RFLP analysis of a PCR product from a variant region of the gene family encoding the major surface glycoprotein (RFLP-MSG) was shown to be discriminative and useful for investigating outbreaks (Ripamonti et al. 2009; Sassi et al. 2012). The polymorphisms in all markers that were used for typing appeared to be stable over months (Keely et al. 1995; Hauser et al. 1997, 2001a; Hauser 2004), which is a prerequisite for microbe-based epidemiological studies (Hauser et al. 1998a). The index of discrimination power is the average probability that two unrelated specimens randomly chosen will be assigned to different types by the method. It has been determined only for the SSCP method and was estimated to be 0.93 (Hauser et al. 2001a), which can be considered as satisfactory because the minimal desired level for a typing system to be used as a single method is 0.95 (Struelens and ESGEM 1996). SSCP and ITSs sequencing identified, respectively, 43 (Hauser 2004) and more than 60 different P. jirovecii genotypes (Lu and Lee 2008). The same markers have been analyzed by SSCP and MLST, but the latter is more discriminative because it detects all polymorphisms.
The interpretation of data using the current typing methods for P. jirovecii must take into account two important caveats. First, co-infection of a single patient with two or more P. jirovecii genotypes has been reported using multiple typing methods (Tsolaki et al. 1996; Hauser et al. 1998a; Nahimana et al. 2000; Ma et al. 2002; Ripamonti et al. 2009), which obviously complicates epidemiological studies (Hauser et al. 1998b). The proportion of co-infected patients varies considerably, ranging from a few percent using direct sequencing to ~70% using SSCP. SSCP can detect a co-infecting type if it represents at least 11% of the population (Nahimana et al. 2000). Cloning of PCR products followed by DNA sequencing can also be used to detect co-infections; the minimal proportion detected will depend on the number of clones that are analyzed. Direct sequencing of PCR products without cloning would not be able to detect low-abundance co-infecting types. Geographical variation or other parameters probably account for some of the reported differences in the proportion of co-infected patients. Identifying all co-infecting genotypes present in clinical specimens of infected humans is probably necessary to fully understand P. jirovecii epidemiology, and may also be useful for analysis of outbreaks, because some genotypes might be more transmissible than others. Nonetheless, at present most investigations of outbreaks use methods that detect only the most abundant genotype(s). Accordingly, recent outbreaks have often been investigated by MLST without cloning to detect co-infections, and this method was able to provide evidence that a single P. jirovecii genotype accounted for all or most of the PCP cases in a given outbreak (Schmoldt et al. 2008; Gianella et al. 2010; Phipps et al. 2011; Pliquett et al. 2012). ITSs sequencing with or without cloning PCR products also reached this conclusion (Yazaki et al. 2009; Le Gal et al. 2012).
The second caveat in interpreting data from typing methods is that the P. jirovecii genotypes present in respiratory samples may not include all those present in the patient’s lungs. Indeed, analysis of lungs from three autopsied individuals in one study demonstrated a compartmentalization of different co-infecting P. jirovecii genotypes in the lungs of two patients. Importantly, some genotypes identified in specific regions of the lungs were absent in the corresponding respiratory specimen (see Table 11.2) (Helweg-Larsen et al. 2001a). As previously stressed by Hughes (2007), this raises questions about studies that concluded that recurrent PCP episodes represent de novo infection, because genotype(s) already present in the lungs may have been detected only during the second and not the first episode. On the other hand, this apparently has not caused problems for investigations of outbreaks since the same P. jirovecii genotype was observed in many instances (see following section).
Table 11.2
Numbers of ITS types corresponding to P. jirovecii genotypes found in various samples of PCP patients by Helweg-Larsen et al. (2001a)
Patient | Respiratory specimena | Autopsy lungs | In common between respiratory specimen and autopsy lungs |
|---|---|---|---|
I | 1 | 1 | 1 |
II | 1 | 3 | 1 |
III | 3 | 10 | 1 |
As far as ITSs typing is concerned, amplification by PCR has been reported to create an artifactual diversity of genotypes, i.e., 37% of them were chimeras (Beser et al. 2007). A substantial decrease in the proportion of these artifacts was obtained by increasing elongation time, primer concentration, and annealing temperature, as well as by decreasing the number of cycles and using a proofreading polymerase (Beser et al. 2007). Nevertheless, two recent studies successfully detected the same ITSs genotype in outbreak PCP cases, apparently without using the advised modifications to decrease chimera formation (Yazaki et al. 2009; Le Gal et al. 2012). Infection with a single genotype may account for this because there would be no opportunity for recombination.
V. Molecular Epidemiology of Pneumocystis Outbreaks
In a recent publication, de Boer et al. (2011) reviewed studies of 16 nosocomial PCP outbreaks among RTRs, six of which used molecular typing to characterize the genotype. A transmission map describing timings of outpatient visits and hospitalizations was generally provided in these studies to assess potential occurrence of infectious encounters (an example is shown in Fig. 11.2). Molecular typing revealed in five out of six instances that all or the vast majority of the outbreak cases with suspected infectious contact(s) harbored the same P. jirovecii genotype, strongly suggesting that inter-human transmission of P. jirovecii had occurred. Importantly, all outbreaks shared the three following features, which probably contributed to the outbreaks: (i) cases received no or inadequate anti-PCP prophylaxis, (ii) contact could occur between potential source patients with clinical or incubating PCP and susceptible patients (in outpatient waiting rooms or during a simultaneous hospitalization), and (iii) no isolation procedure of PCP patients was utilized. However, being retrospective, these studies could not firmly exclude that cases acquired the fungus from asymptomatic colonized health-care worker(s), from an unidentified environmental source, or by indirect transmission from an index patient through nosocomial carriers.
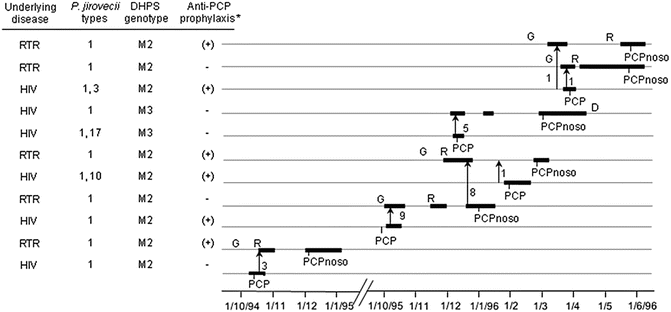

Fig. 11.2
Potential encounters compatible with nosocomial inter-human transmission of P. jirovecii in one building of a hospital. Thicker parts of solid lines represent periods of hospitalization. Each encounter or consecutive encounters are figured by an arrow pointing to the direction of the presumed transmission; the number of encounters is indicated close to each arrow. *Anti-PCP prophylaxis with Fansidar was suboptimal (25 mg pyrimethamine and 500 mg sulfadoxine once or twice every 2 weeks). D death, G graft, R rejection episode, RTR renal transplant recipient, PCPnoso nosocomial case, DHPS dihydropteroate synthase, M2 amino acid change within the putative sulfa-binding site at position 57 (Pro to Ser) [Taken from Fig. 4 of Rabodonirina et al. (2004)]
Since the review of de Boer et al. (2011), eight additional PCP outbreaks have been reported in seven studies, nearly all in RTRs. One study provided evidence of transmission only on the basis of possible infectious encounters (Mori et al. 2010). Of note, the latter is the first among outpatients with rheumatoid arthritis. In addition to possible infectious encounters, the other six studies reported that a single molecular P. jirovecii genotype accounted for most or all of the RTR outbreak cases (Phipps et al. 2011; Thomas et al. 2011; Wynckel et al. 2011; Brunot et al. 2012; Pliquett et al. 2012; Le Gal et al. 2012). In all eight outbreaks, patients were not receiving anti-PCP prophylaxis at the time of suspected exposure, and there was no policy to isolate PCP patients. Thus, these recent studies provide further evidence that inter-human transmission is involved in such outbreaks, but they also could not exclude a common source or indirect transmission.
These studies highlight at least three new issues that could help explain PCP outbreaks:
(i)
For the first time, three out of ten potential source patients were identified as carriers of P. jirovecii without clinical symptoms of PCP (Le Gal et al. 2012). This is important because carriers are not routinely identified within hospitals and they may be even more numerous than PCP patients, and as such they may constitute an important nosocomial source of P. jirovecii. The risk of transmission by such individuals needs to be determined, potentially by measuring dissemination of the fungus in the air as previously described for PCP patients (Choukri et al. 2010). If such individuals are an important source of infection, approaches to minimizing such transmission would need to be evaluated.
(ii)
The new studies identified new clinical risk factors for PCP that may have played a role in the outbreaks. Alteration in immunosuppressive therapy, graft rejection, cytomegalovirus infection, and older age were previously identified (de Boer et al. 2011). A cohort analysis including 14 cases and 324 control patients identified two new independent risk factors: underlying pulmonary disease and transplanted organ dysfunction (Phipps et al. 2011). Lower CD4 lymphocyte counts at 3 months after transplantation (Brunot et al. 2012) or just before PCP diagnosis (Struijk et al. 2011) were also identified as risk factors. Although they need confirmation because they were identified in single studies, these findings may be important for preventing outbreaks by better identifying patients who would benefit from PCP prophylaxis.
(iii)
Two different P. jirovecii genotypes were involved in two outbreaks that occurred at a relatively close geographical distance of 50 km (Liverpool and Manchester) (Thomas et al. 2011). This raises new issues for the analysis of the different outbreak-associated P. jirovecii genotypes, which we present in the following section.
VI. P. jirovecii Outbreak Genotypes
An important finding concerning P. jirovecii genotypes involved in outbreaks has been reported recently (Sassi et al. 2012). Genotypes linked to two PCP outbreaks that occurred contemporaneously in two European cities at a distance of 300 km (Zurich and Munich) were found to be identical using both MLST and RFLP-MSG. Subsequently, the same genotype was identified using MLST in a third contemporaneous outbreak at a distance of 400 km from the two previous ones (Frankfurt) (Hauser et al. 2013). However, this genotype was different from that involved in a contemporaneous Japanese outbreak (Nagoya) (Sassi et al. 2012), as well as from that observed in another European outbreak that occurred about 12 years earlier at a distance of 400–800 km (Lyon) (Hauser et al. 2013).
Two epidemiological factors may have played a role in the variation of the genotype involved in the different outbreaks: the period of time and the geographical location (Hauser et al. 2013). However, the presence of the same genotype in three contemporaneous and geographically close outbreaks is particularly intriguing. As previously noted (Sassi et al. 2012; de Boer 2012), two hypotheses could explain this latter finding: (i) the genotype was predominant in an area covering the three European locations and infected index patients of the three outbreaks, or (ii) the genotype carried uncharacterized pathogenic factor(s) and some unidentified contact (e.g., a patient or colonized individual) existed between the three outbreaks. Because the few sporadic contemporaneous cases analyzed in the three outbreaks (six, three, and two in Munich, Zurich, and Frankfurt, respectively) were infected by other genotypes, the outbreak genotype was considered not to be predominant, but was hypothesized to be more pathogenic in RTRs (Sassi et al. 2012).
On the other hand, by analysis of a large number of isolates, the Lyon outbreak genotype was demonstrated to be the predominant one at the time in the affected hospital as well as in the local area (Rabodonirina et al. 2004), and one of the most prevalent in several European locations (Hauser et al. 2001a). Nevertheless, the Lyon genotype may have been not only predominant but also more transmissible. Indeed, the potential infectious encounters during the outbreak also involved other genotypes, but these apparently were not transmitted (Rabodonirina et al. 2004). The increased transmissibility or pathogenicity of the Lyon genotype might have been due to unidentified specific factors, but also potentially to the presence of one or two amino acid changes within the putative sulfa-binding site of P. jirovecii dihydropteroate synthase (Pro to Ser at position 57, mutation M2; Thr to Ala and Pro to Ser at positions 55 and 57, respectively, double mutation M3). All but one potential donor in the encounters harbored M2, whereas M3 was present in the remaining donor (Fig. 11.2). Epidemiological studies have shown that these mutations confer some level of resistance to sulfonamides (Kazanjian et al. 1998; Helweg-Larsen et al. 1999; Nahimana et al. 2003a), and the results were confirmed by functional complementation in Escherichia coli (Iliades et al. 2005) and Saccharomyces cerevisiae (Meneau et al. 2004). Consequently, the Lyon outbreak genotype may have been selected among the different genotypes encountered because several patients were receiving suboptimal prophylaxis (Fig. 11.2). Thus, the observations in Lyon suggest that the two hypotheses, preponderance and increased pathogenicity of the outbreak genotypes, may not be mutually exclusive.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


