(1)
The Molecular Cardiology and Neuromuscular Institute, Highland Park, NJ, USA
Abstract
Cardiac mitochondria are complex highly organized cellular organelles, which play central roles not only in energy homeostasis but also in various biosynthetic, signaling, and cell death pathways. A wide range of methodological approaches have been developed to assess mitochondrial function in the heart. High- and super-resolution fluorescent microscopy has been used to visualize mitochondria and measure number of mitochondrial characteristics. Electron microscopy and electron tomography can visualize not only mitochondrial ultrastructure but also mitochondrial multiprotein complexes at near-atomic resolution. A whole arsenal of modern molecular biological methods has been exploited in analysis of mtDNA and its dynamics. The in vitro spectrophotometric enzyme assays and polarographic measurements of oxygen consumption are commonly utilized to assess mitochondrial function. Noninvasive methods based on magnetic resonance spectroscopy (MRS) have emerged as a powerful tool to study mitochondrial function in vivo in human heart. Electrophoretic techniques, such as 1D-, 2D-PAGE, and BN-PAGE, have proved to be a very sensitive and informative approach to analyze complex content of mitochondrial proteins. Advances in separation and mass spectrometry (MS)-based technologies have led to the identification of a significant number of mitochondrial proteins from various rodent and human tissues, including heart. Animal models are of a great utility for the investigation of mitochondrial functions and their roles in the heart; however, mtDNA gene targeting still presents a significant technical challenge. Great advances in these methodological approaches have fueled progress in our understanding of mitochondrial functional role in heart physiology and pathophysiology.
Introduction
Cardiac mitochondria are complex highly organized cellular organelles, which play central roles not only in energy homeostasis but also in various biosynthetic, signaling, and cell death pathways. Moreover, mitochondria are highly dynamic organelles that continuously divide and fuse as well as move within the cell. Therefore, it is not surprising that multiple methodological approaches have been developed to assess mitochondrial functions.
Since significant difficulties with obtaining samples of the human heart exist, traditionally the majority of the studies on the roles of mitochondria in cardiac physiology and pathophysiology have been performed on animal models. However, recent advances in the development of more sensitive methods to analyze mitochondria make possible to use smaller amount of cardiac tissue available from heart surgeries. Also, human cardiomyocytes derived from either neonates or adults and cultured in vitro have proved to be a highly informative model to study human cardiac mitochondrial functions.
In this chapter, we will discuss major cytochemical, molecular biological, and biochemical techniques exploited to investigate cardiac mitochondria. We will focus especially on recent developments in technologies assessing mitochondrial function.
High-Resolution Imaging of Mitochondria in Live Cells
Given the difficulties to visualize mitochondria using various phase contrast or interference contrast optics, in the last decades, most studies on mitochondrial morphology and dynamics have relied on far-field fluorescence microscopy [1, 2]. This approach relies on the development of microscopic techniques and specific fluorescent probes to stain mitochondria or to label individual mitochondrial proteins. Recent advances in fluorescent imaging technologies have significantly enhanced our ability to analyze mitochondrial morphology and dynamics and precisely measure levels of specific metabolites and ions within its sub-compartments, such as the mitochondrial membranes and matrix.
A variety of fluorescent probes and potentiometric dyes listed in Table 2.1 have been increasingly used to quantitatively evaluate overall cardiomyocyte mitochondrial number, membrane potential, oxidative stress, apoptosis, and Ca2+ concentrations [3–6]. The uptake of the majority of these dyes into mitochondria depends on the mitochondrial membrane potential. The fluorescence of some of these dyes changes depending on the environment and can be used to measure the mitochondrial membrane potential.
Table 2.1
Fluorescent dyes to study mitochondria
Fluorescent dyes | Fluorescence maximum (nm) | Comments | References | |
|---|---|---|---|---|
Excitation | Emission | |||
3,3′-Dihexyloxacarbocyanine iodide (DiOC6) | 488 | 501 | Can be used as an indicator of the mitochondrial membrane potential ΔΨm; may inhibit mitochondrial respiration | [7] |
5,5′,6,6′-Tetrachloro-1,1′,3,3′-tetraethylbenzimidazolylcarbocyanine iodide (JC-1) | 514 | 527 and 590 | Can be used as an indicator of ΔΨm; emission spectral shift depends on dye concentration | |
MitoTracker MitoFluor | Several mitochondrial dyes with different characteristics | [5] | ||
Nonyl acridine orange (NAO) | 495 | 522 | Uptake does not depend on ΔΨm | [9] |
Rhodamine 123 (Rhod 123) | 507 | 529 | Can be used as an indicator of ΔΨm; may inhibit mitochondrial respiration | [4] |
Tetramethylrhodamine ethyl ester (TMRE) Tetramethylrhodamine methyl ester (TMRM) | 549 (548) | 574 (573) | Both dyes are similar to rhod 123, but more membrane permeable; can be used as an indicator of ΔΨm; may inhibit mitochondrial respiration | |
While these fluorescent dye markers stain the whole organelle, the discovery and cloning of the green fluorescent protein (GFP) from the jellyfish Aequorea victoria in the early 1990s made possible to analyze the dynamics of specific mitochondrial proteins. Shortly after GFP cloning and heterologous expression, it has been used in mitochondrial studies in the living cells [11]. Since then, a variety of fluorescent proteins (FPs) as a toolkit for in vivo imaging has been produced [12–14]. Based on various FP variants, molecular sensors to measure a number of mitochondrial parameters, such as redox potential, Ca2+ and Cl− levels, and pH, are currently available [15–20]. A complementary strategy using fusion proteins labeled with synthetic fluorescent molecules has also been suggested; however, so far it has not been exploited for the visualization of mitochondrial proteins [21, 22].
Recently, “nanoscopy” or “super-resolution” fluorescence technologies have been introduced to overcome the limiting role of diffraction in a lens-based optical microscopy and to provide nanometer-level precision coordinates [2, 23–25]. To this end, several physical concepts relaying on reversible saturable optical fluorophore transitions have been developed, such as stimulated emission depletion microscopy and ground state depletion microscopy [26, 27]. Using these technologies, it has recently been demonstrated the distribution of various proteins in mitochondria and the flow of the mitochondrial inner membrane in live cells with a nanoscale resolution [28–30].
High-Resolution Electron Microscopy and Electron Tomography
In 1953, Palade and Sjostrand published pioneering electron micrographs showing mitochondria as double-membrane enclosed organelles with an inner membrane forming numerous invaginations, cristae [31, 32]. Since then, electron microscopy (EM) has tremendously advanced and during the last six decades has become a powerful tool to study mitochondrial ultrastructure and function. Various modifications of the fixation, dehydration, sectioning, and staining of section better preserving the native mitochondrial morphology have been developed. Combination of EM with immunostaining techniques has significantly enhanced the power of this technique and has provided further insights into the mitochondrial architecture and function.
Since conventional transmission EM generates two-dimensional images of three-dimensional (3D) objects, 3D imaging techniques such as high-resolution scanning electron microscopy and electron tomography (ET) have been introduced (Fig. 2.1). This revolutionizing approach is able to yield 3D reconstruction of mitochondria at molecular levels [33–37].
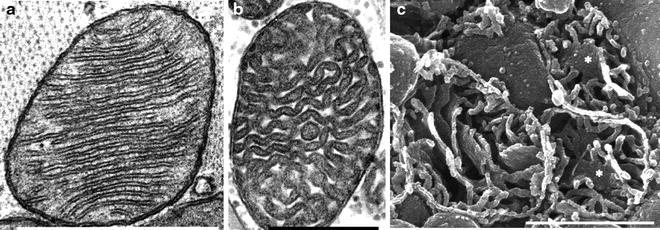

Fig. 2.1
Transmission electron microscopy (a, b) and scanning electron microscopy (c) of cardiac mitochondria. (a) Rat cardiac mitochondrion with lamelliform cristae. (b) Human cardiac mitochondrion with tubular cristae; the tissue sample was from the right side of the interventricular septum. (c) Three transected human cardiac mitochondria prepared by the osmium-extraction technique are shown. The scale bar is 0.5 mm (a, c) or 1 mm (b) (adapted from Hoppel et al. [38] with permission of Elsevier)
The newly emerging cryo-ET using quickly frozen samples is devoid of artifacts induced by chemical fixation, dehydration, and staining [38–41]. Cryo-ET, combined with 3D image classification and single particle averaging, can visualize not only mitochondrial ultrastructure but also mitochondrial multiprotein complexes at near-atomic resolution (Fig. 2.2) [42–44].
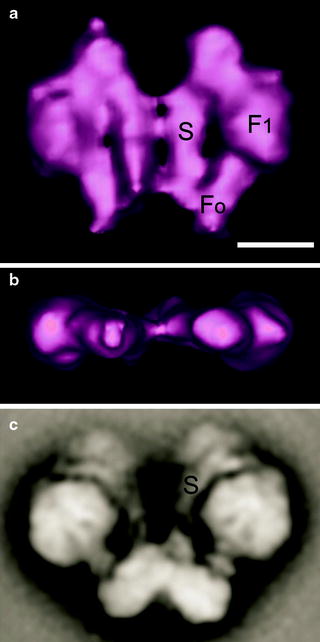

Fig. 2.2
Electron tomography and single particle electron microscopy of dimeric ATP synthase from Polytomella mitochondria. (a) Average of tomographic sub-volumes of ATP synthase dimer seen from a side revealing the F1 headpiece, the Fo membrane part, and the peripheral stalk (S) or stator. (b) Same average, seen from the top. (c) Average from negatively stained side views of isolated dimeric ATP synthase. The scale bar is 10 nm (adapted from Dudkina et al. [44] with permission from Elsevier)
Molecular Biological and Biochemical Methods
Most of molecular and biochemical studies of cardiac mitochondria have relied on isolated organelles. In the past decades, numerous isolation and fractionation procedures of mitochondria as well as their membrane-bound sub-compartments together with the identification of specific markers have been reported. The major drawback in study of human cardiac mitochondria is the difficulty in obtaining sufficient amount of fresh tissue, especially from control healthy human heart.
Isolated cardiac mitochondria retain their essential morphological and functional characteristics. If a substantial amount of tissue is available (e.g., from the heart of patients undergoing transplantation or during routine cardiac surgery), two characteristic cardiac populations the subsarcolemmal mitochondria (SSM) and interfibrillar mitochondria (IFM) can be isolated [45–47]. Isolated SSM and IFM have characteristic lamelliform and tubular cristae, respectively [35].
With permeabilized myocardial fibers, the major problem of lack of sufficient amount of tissue can be overcome [48, 49]. This method allows studying cardiac mitochondria in their cellular environment using only a few milligrams of tissue. Both SSM and IFM populations have been detected and evaluated by the permeabilized fiber method [48]. Importantly, like with isolated mitochondria, various substrates, activators, and inhibitors can be used to analyze mitochondrial function.
mtDNA Analysis
Since mitochondria have their own DNA (mtDNA), a whole arsenal of modern molecular biological methods have been exploited in analysis of mtDNA and its dynamics. They include a variety of amplification and mutation detection techniques to screen for maternally inherited mtDNA point mutations and large-scale mtDNA deletions, Southern and Northern blotting, accessing mitochondrial copy number as well as improved techniques for the analysis of mtDNA damage and repair. Several excellent books containing updated methods are currently available.
The development of cultured mammalian cells, which lack mtDNA, due to growth in low concentrations of ethidium bromide, was pivotal to study the effect of specific mutations on mitochondrial function [50, 51]. These cells similar to yeast petite mutants, which lack mtDNA, are termed rho 0 cells. They exhibit defective respiration and adopt an anaerobic phenotype. Cytoplasts containing mitochondria can be prepared from a wide variety of enucleated cells (e.g., platelets, fibroblasts) and fused with rho 0 cells lacking mtDNA to form cell hybrids (cybrids), essentially changing the nucleus-mitochondria content. Cybrids containing normal mitochondria regain functional respiration, manifest an aerobic phenotype, and can be readily distinguished from cybrids with defective mitochondria. Cybrids can be maintained in culture using the appropriate media supplementation and have been successfully employed to study nuclear-mitochondrial interactions, as well as the effects of specific mitochondrial mutations in different nuclear backgrounds.
In Vitro Assessment of Mitochondrial Function
Mitochondria produce energy required for the rhythmic contraction of the heart by two main metabolic pathways—glycolysis and oxidative phosphorylation (OXPHOS) that couples the oxidation to phosphorylation of ADP to ATP. Mitochondria produce approximately 90% of cellular ATP needed to drive numerous energy-requiring processes. Mitochondrial dysfunction related to different cardiac pathologies can be caused by the impairments of any step of this very complex multistep process. Furthermore, such alterations may be related to the quantity, functionality, or interactions between the numerous components of the process.
Several in vitro and in vivo techniques for assessing mitochondrial function have been developed (Fig. 2.3) [52–54]. The in vitro measurement of activities of various mitochondrial enzymes is commonly used to estimate the functionality of specific steps implicated in mitochondrial metabolism. A comprehensive list of mitochondrial enzyme activities associated with various pathologies in the human heart has been summarized in the recent review of Lemieux and Hoppel [55]. Spectrophotometric-based enzyme assays require small amounts of tissues and therefore are well suited for human studies. Due to the limiting amount of human cardiac tissue, most of the studies have been performed on tissue homogenates rather than on isolated mitochondria.
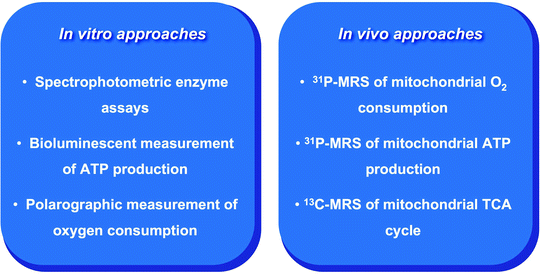

Fig. 2.3
Main in vitro and in vivo approaches for the assessment of mitochondrial function. See text for the details. MRS magnetic resonance spectroscopy; TCA tricarboxylic acid
Although the measurements of individual mitochondrial enzyme activities provide valuable insights into mitochondrial function, they cannot accurately reflect the integral mitochondrial function. Two major in vitro approaches serve this purpose: a bioluminescent measurement of ATP production and polarographic measurement of oxygen consumption. Oxidation of the substrate luciferin catalyzed by firefly luciferase in an ATP-dependent manner generates the light signal, which is proportional of ATP concentrations [52, 56, 57]. Light emission can be precisely measured by a luminometer to quantify the rates of ATP production. The combination of various substrates allows to selectively assess different mitochondrial complexes. Fresh cardiac tissue has to be used for the isolation mitochondria since freezing-thawing disrupts the membrane structure.
Polarographic measurement of oxygen consumption in mitochondria in the presence of specific substrates represents an alternative in vitro approach to assess mitochondrial functional activity (OXPHOS) [58]. Although conventional respirometry has required large amounts of tissue for accurate measurements, the development of high-resolution respirometry allows measurements of oxygen consumption in very small biopsy samples [59–61]. Both very small amounts of isolated mitochondria (0.01 mg) and permeabilized muscle fibers can be used. In addition, serial measurements in the same sample are possible. Important advantage of in vitro approaches is their ability to use various substrates, cofactors, activators, or inhibitors/uncouplers.
In Vivo Assessment of Mitochondrial Function
Noninvasive methods based on magnetic resonance spectroscopy (MRS) have emerged as a powerful technology to study mitochondrial function in vivo in various human tissues including the heart [53, 62–65]. MRS measures magnetic resonance signals from MR visible nuclei, such as 13carbon (13C), 1hydrogen (1H), 31phosphorus (31P), and 23sodium (23Na). The radiofrequency (RF) generator of a spectrometer produces an RF impulse to excite the nuclear spins in the myocardium of the subject. The resulting magnetic resonance signal, free induction decay, is recorded and mathematically processed to yield a magnetic resonance spectrum (Fig. 2.4).
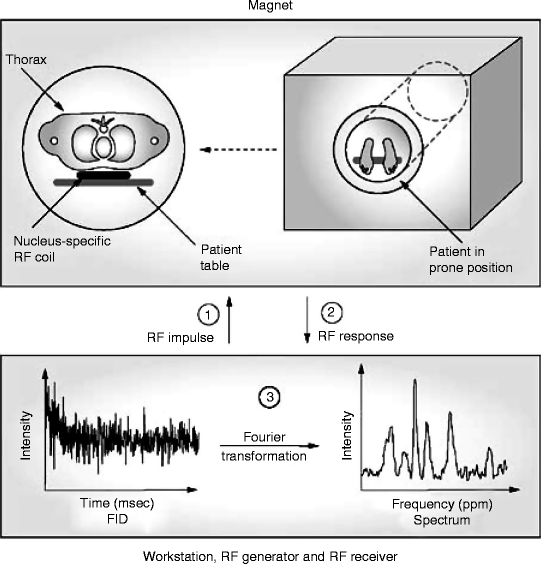

Fig. 2.4
Scheme of a human cardiac MRS analysis. See text for details. FID free induction decay; MRS magnetic resonance spectroscopy; RF radiofrequency
A typical 31P spectrum obtained from a healthy subject consists of 6 frequency resonances: α-, β-, and γ- 31P atoms of ATP; phosphocreatine (PCr); 2,3-diphosphoglycerate (from erythrocytes); and phosphodiesters (from cell membranes and serum phospholipids) (Fig. 2.5) [66]. Thus, each peak at specific resonant frequency, also called a chemical shift, expressed in parts per million (ppm) along the x-axis, corresponds to the specific metabolite, while the peak amplitude along the y-axis corresponds to the metabolite concentration.
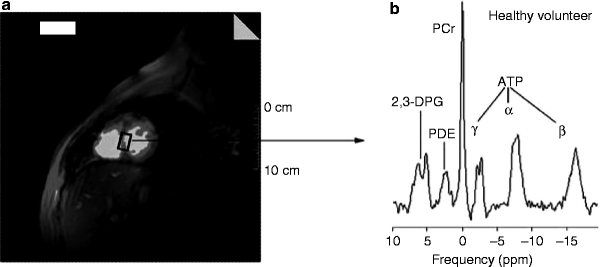

Fig. 2.5
31P-MRS of a healthy subject. (a) 1Hydrogen short-axis scout image showing the voxel selection in the myocardial interventricular septum of a healthy subject. (b) A typical human cardiac 31P spectrum from a healthy subject showing the following six resonances: three 31P atoms of ATP (α, β, and γ); PCr; 2,3-DPG; and PDE. 2,3-DPG 2,3-diphosphoglycerate; PCr phosphocreatine; PDE phosphodiesters (adapted from Hudsmith and Neubauer [64] with permission from Nature Publishing Group)
This noninvasive technique allows monitoring mitochondrial energy metabolism. 31P-MRS enables in vivo detection of ATP and PCr dynamics, whereas 13C-MRS assesses the tricarboxylic acid (TCA) cycle, glycolysis, or β-oxidation. The PCr/ATP ratio, most commonly determined by cardiac 31P-MRS, is an important indicator of the energetic state of the myocardium. In ischemia, demand for ATP outweighs its production leading to decrease in the PCr levels and therefore to a low PCr/ATP ratio [67]. In heart failure, low PCr/ATP ratio can also result from a reduction of the total creatine pool correlating with the severity of the condition [68–70].
Measurements of mitochondrial ATP production assessed by 31P-MRS combined with the 13C-MRS measurements of oxidation via the TCA cycle allow to evaluate in vivo mitochondrial coupling between oxidation and ATP synthesis [53, 71].
MRS technologies have proved to be a useful in vivo approach for characterization of myocardial energetics in the experimental setting. However, rather poor reproducibility, low spatial and temporal resolution, and long acquisition times limit currently their clinical application.
Electrophoretic Techniques to Study Mitochondria
One- and more recently introduced two-dimensional polyacrylamide gel electrophoresis (1D- and 2D-PAGE, respectively) followed by Western immunoblotting has proved to be a very sensitive and informative approach to analyze the complex content of mitochondrial proteins [72–74]. While 1D-PAGE separates proteins by their mobility, which mainly depends on their molecular mass, 2D-PAGE separates proteins based on their two characteristics, isoelectric point (pI) and molecular mass. This results in highly increased resolution and thereby better identification of proteins. However, hydrophobic mitochondrial membrane proteins create significant problems for their separation, due to their low solubility, leading to their underrepresentation in the 2D-PAGE analyses [75]. Modifications of traditional 2D-PAGE have been developed to overcome this shortcoming and improve the separation of mitochondrial membrane proteins [76, 77].
In addition to conventional 1D- and 2D-PAGE, blue-native PAGE (BN-PAGE) was developed to fractionate large mitochondrial multiprotein complexes [78–80]. This technique can efficiently separate hydrophobic mitochondrial membrane proteins and protein complexes in the mass range of 10 kDa to 10 MDa. In addition to native molecular size, it also enables to determine protein composition, stoichiometry, and relative abundance of mitochondrial multiprotein complexes (Fig. 2.6).
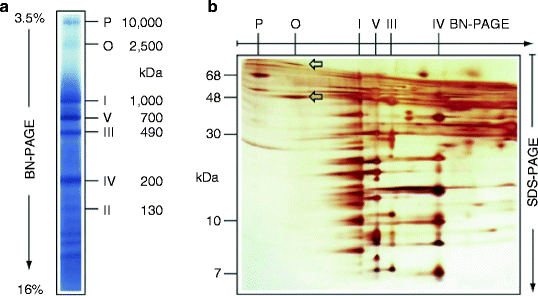

Fig. 2.6
Separation of dodecylmaltoside-solubilized mitochondrial complexes by blue-native polyacrylamide gel electrophoresis (the native mass range <100 kDa to ∼10 MDa). (a) Bovine cardiac mitochondrial complexes I–V, pyruvate dehydrogenase complex (P), and oxoglutarate dehydrogenase complex (O) were separated according to their indicated masses by blue-native polyacrylamide gel electrophoresis (BN-PAGE) in a linear 3.5–16% polyacrylamide gradient gel. (b) For the second dimension, subunits of the native complexes were resolved into individual subunits by tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The 96-kDa and 48-kDa subunits of the oxoglutarate dehydrogenase complex are indicated by arrows (adapted from Wittig et al. [80] with permission from Nature Publishing Group)
Depending on the stability of the protein complexes of interest, various nonionic detergents can be used for solubilization of mitochondrial membranes. The anionic stain Coomassie blue G-250, added to the solubilized sample, binds to membrane proteins imposing negative charge on the proteins. As a result, even basic proteins at neutral pH migrate through the gel to the anode according to their molecular sizes in the gradient BN-PAGE (Fig. 2.6a). Importantly, separated proteins or complexes remain native; therefore, various in-gel activity assays can be performed or blue protein bands can be eluted from the gel and used for further characterization [81–83].
The second dimension of BN-PAGE under stronger detergent conditions can be applied to resolved supramolecular assemblies into the distinct complexes and interacting partners [82, 84–86]. Finally, the complexes separated by BN-PAGE can be resolved into their individual subunits by applying SDS-PAGE for the second dimension (Fig. 2.6b).
This approach has been applied to identify and quantitate OXPHOS proteins, to demonstrate the dimerization of yeast mitochondrial F1F0-ATP synthase and the formation of a mitochondrial supercomplex by respiratory-chain complexes I and III [87–90]. Moreover, BN-PAGE has been used to monitor the assembly of voltage-dependent anion channel (VDAC) and the translocase complex in the mitochondrial outer membrane [91–93].
Described above electrophoretic techniques can be implicated as separation steps in Western immunoblot or proteomic analysis of mitochondrial proteins. Highly specific and sensitive immunodetection methods significantly enhance the power of electrophoresis techniques. A growing list of specific antibodies to various mitochondrial proteins has been used in both immunocytochemical and Western immunoblot analysis. Remarkably, an increasing number of these antibodies (e.g., ATP synthase subunits, cytochrome c and cytochrome c oxidase, PGC-1α, mtTFA) are now commercially available from companies such as MitoSciences (Eugene, Oregon), Santa Cruz Biotechnology (Santa Cruz, CA), BD Pharmingen (San Diego, CA), Millipore (Billerica, MA), Affinity Bioreagents (Golden, CO), and Abcam (Cambridge, MA).
Mitochondrial Proteomics
The completion of the Human Genome Project has highlighted the crucial importance of functional proteomic studies, which focus on identification, quantification, modification, and localization of cellular proteins. Computational analysis of the human genome has predicted that the human mitochondrial proteome consists of approximately 1,500 distinct proteins [94–98]. However, the identity and especially functional characteristics of many of them remain largely unknown.
Mass spectrometry (MS)-based technologies represent the most comprehensive and versatile tool in large-scale proteomics. The MS-based mitochondrial proteome analysis includes several steps [99]:
Purification of mitochondria
Solubilization and digestion of mitochondrial proteins
Analysis of the digested peptides by various modifications of MS
Identification of the obtained peptide sequences using protein databases
While crude mitochondrial protein can be used for MS analysis, introduction of purification/fractionation steps, such as 1D-PAGE, 2D-PAGE, BN-PAGE, or liquid chromatography, prior to MS analysis increases chances that low abundant proteins will be identified (Fig. 2.7). In addition, these purification steps provide important physicochemical information (e.g., MW and pI) of a protein. Moreover, different mitochondrial compartments, such as the outer and inner membranes or matrix, can be purified and used for MS analysis [100].
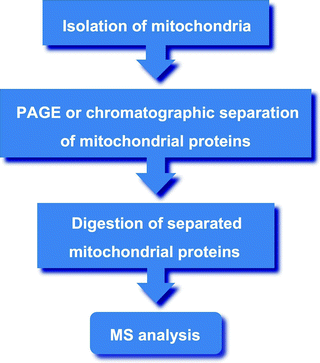

Fig. 2.7
Major steps of the mass spectrometry (MS)-based mitochondrial proteome analysis. See text for details. PAGE, polyacrylamide gel electrophoresis
Affinity purification techniques to enrich crude mitochondrial proteins for Ca2+ binding, hydrophobic proteins, or glycoproteins have been reported [101, 102]. Immuno-precipitation can be used to purify multiprotein mitochondrial complexes and therefore increase the chances to identify their subunits or interacting proteins [103]. Given that posttranslation modifications (e.g., acetylation, nitration, and phosphorylation) of mitochondrial proteins play important regulatory roles, immunoaffinity purification can also be used to purify the modified mitochondrial proteins prior to MS [104].
Pioneering proteomic studies in 1998 and 2001 were performed on isolated human placental and human neuroblastoma mitochondria using 2D-PAGE followed by peptide mass fingerprinting or MS [105, 106]. 46 and 80 mitochondrial proteins were identified, respectively. Advances in separation and MS technologies have enabled detection of low abundant proteins and led to the identification of a significant number of mitochondrial proteins from various rodent and human tissues, including the heart [74, 96, 107–110]. For example, Pagliarini et al. [111] have recently identified 3,881distinct proteins in the mitochondrial preparations isolated from 14 different mouse tissues.
Notably, not all proteins identified in isolated mitochondria preparations are bona fide mitochondrial proteins; many of them represent contaminants of the preparations. High sensitivity of novel technologies highlights the importance to distinguish contaminants using the highly purified mitochondrial preparation as well as computational approaches [98]. On the other hand, careful analysis has suggested that approximately 15% of mitochondrial proteins can be localized to multiple cellular compartments [109, 112].
Mootha and coworkers [113] have suggested an integrative genomic approach to expand the catalog of human mitochondrial proteins. As a result of these efforts, the comprehensive mitochondrial catalog, MitoCarta (http://broadinstitute.org/pubs/MitoCarta/), containing 1,098 different gene loci, has been created. Currently, six online databases of mitochondrial proteins are available, including, in addition to MitoCarta, MitoProteome (http://mitoproteome.org), MitoP2 (http://ihg.gsf.de/mitop2), MitoMiner (http://mitominer.mrc-mbu.cam.ac.uk), MitoPhenome (http://mitophenome.org), and HMPDb (http://bioinfo.nist.gov) [114–117].
Finally, the impressive progress achieved in our understanding of cardiac mitochondria function and in computational analysis results in the development of mathematical models of these organelles. Modeling of two aspects of mitochondrial metabolism, bioenergetics and Ca2+ dynamics, has been especially productive [118–126]. Although this approach has shown its potential, more work is needed to increase the predictive accuracy of mathematical models of cardiac mitochondria.
Transgenic Models for Assessing Mitochondrial Function in the Heart
Animal transgenic models are of a great utility for the investigation of mitochondrial functions and their roles in heart physiology and pathophysiology. Little information is currently available concerning mtDNA gene targeting (i.e., generation of null mutations or gene knockouts) since generation of mtDNA gene knockouts presents a significant technical challenge. However, there is a growing list of murine models harboring a relatively wide spectrum of targeted nuclear genes encoding mitochondrial proteins. Importantly, many of these mitochondrial alterations are associated with cardiac dysfunction resembling various human cardiac phenotypes.
Disruption of genes for the factors involved in mitochondrial energy production, the adenine nucleotide translocase (ANT), Mn2+ superoxide dismutase (MnSOD), regulators of lipid metabolism (peroxisome proliferator-activated receptors (PPARs)) mitochondrial trifunctional protein (MTP) subunits) and several transcription factors has been reported [55, 127–132]. The majority of those transcription factors/regulators including estrogen-related receptors (ERRs), GA-binding protein (GABD, also referred to as NRF-2), nuclear respiratory factror-1 (NRF-1), members of PPAR-γ coactivator-1 (PGC-1) family, and mitochondrial transcription factor A (TFAM; also termed mtTFA) are involved in mitochondrial biogenesis [131–134]. Notably, impairments of these factors not only result in mitochondrial dysfunction but can also lead to severe cardiomyopathy. The use of cardiac-specific knockout or overexpression of some of these regulators has also been informative in our understanding of the role of mitochondria in cardiac dysfunction [135]. Importantly, the development of animal models for mitochondrial-based cardiac dysfunction also offers the opportunity of testing novel therapeutic strategies for the treatment of cardiovascular diseases (CVDs).
Although animal models have provided important insights into mitochondrial function, the particularities of the human cardiac mitochondria have to be taken into account. For example, the OXPHOS capacity in the human heart is comparable to that of skeletal muscle, whereas, in rodents, the OXPHOS capacity in the heart is much higher than in skeletal muscle [136, 137]. Significant differences in Ca2+ homeostasis between human and animal cardiac mitochondria have also been reported [138].
Conclusion and Future Progress
Great advances in a variety of experimental technologies have fueled progress in the understanding of mitochondrial functional role in heart physiology and pathophysiology. Cardiac mitochondria have been studied extensively using traditional histological, biochemical, molecular, and genetical approaches as well as newly developed technologies.
Conventional light microscopy has been used for decades to study mitochondria in various human tissues, including the heart. The development of microscopic instrumentations in the last decades and the generation of various fluorescence stains to label mitochondria on the whole and their individual proteins have made possible to study these organelles in live cells in real time. Mitochondria imaging in the living cells has further been advanced by the discovery of various FPs and progress in molecular cloning strategies. Currently many FP-tagged mitochondrial proteins have been genetically engineered and microscopically identified to be specifically localized to mitochondria. An alternative strategy based on a tag-mediated labeling of mitochondrial proteins using synthetic fluorescent molecules has to be tested in mitochondrial studies. Importantly, both tag-mediated labeling approaches allow to not only localize the organelle or its components but also to monitor essential functional parameters, such as mitochondrial membrane potential, pH, Ca2+, or Cl– levels. The generation of different fluorescent probes to monitor additional parameters is in progress.
Progress in wide-field fluorescence and confocal laser-scanning microscopic techniques allows the recording of mitochondrial dynamics in three spatial dimensions over time, 4D imaging [139]. Although a few limitations remain, time-lapse microscopy is a powerful technique to study mitochondrial dynamics in the heart during various physiological and pathological conditions.
In the past decade, fluorescence resonance energy transfer (FRET) microscopy has emerged as a very powerful tool to study protein-protein interactions in the living cells [140]. This approach is relied on the use of proteins tagged with different fluorophores. FRET is an energy transfer from an excited donor fluorophore to a nearby, at a distance of 1–10 nm, acceptor fluorophore. FRET has been used to study dynamic interactions of pro-apoptotic proteins, subunits of the F1F0-ATP synthase complex, and Ca2+ dynamics within mitochondria [141–145].
Recently, revolutionizing super-resolution fluorescence microscopy, also termed nanoscopy, has emerged enabling the recording of mitochondrial protein complexes with a 3D nanoscale resolution.
Ongoing progress in EM, high-resolution scanning EM and ET allows 3D visualization of mitochondria and mitochondrial complexes at near-atomic resolution. Remarkably, these techniques can be applied on nonchemically fixed, unstained, frozen samples, which preserve better their native characteristics. Given recent improvements, the resolution may reach 2 nm, which is close to the theoretical limit [44, 146].
Many experimental approaches to study mitochondria, histochemical as well as molecular and biochemical, rely on the use of antibodies against various mitochondrial markers. Although their list is continuously growing, the generation of highly specific antibodies represents an important task in the field. Monoclonal antibodies directed against distinct epitopes of mitochondrial proteins are particularly valuable to get insights into the molecular mechanisms of mitochondrial function.
Human mtDNA, which contains 37 genes including 13 genes for subunits of OXPHOS complexes, 2 genes for ribosomal RNAs (rRNAs), and 22 for transfer RNAs (tRNAs), has been extensively sequenced across populations. The rest of approximately 1,500 mitochondrial proteins are encoded by the nuclear genome. Due to the generation of reactive oxygen species (ROS) in mitochondria and the lack of histones, mtDNA is highly susceptible to oxidative as well as other types of DNA damage. Multiple mutations in both nuclear and mitochondrial genes have been identified to be associated with mitochondrial disorders [147–149]. Despite significant progress in deciphering the molecular mechanisms of highly heterogeneous mitochondrial diseases, it is still unclear how certain mutations are translated into a broad range of clinical phenotypes.
One of the major obstacles to functional studies of mtDNA defects results from the lack of an effective technique of targeting mtDNA in mammalian cells. Several alternative approaches utilizing an antisense targeting, signal peptide-targeting sequences covalently attached to oligonucleotides, mitochondria-targeted restriction, or Zn-finger nucleases have been suggested [150]. Moreover, since the current diagnostics of mitochondrial disorders requires the use of multiple techniques and the development of a novel one-step, nuclear, and mitochondrial genome, comprehensive analysis is highly desirable [151].
Recent impressive progress in high-resolution respirometry, allowing measurements of oxidative capacity in very small samples, and in bioluminescent measurements of ATP production has led to appreciation of these methods as a “gold standard” for in vitro mitochondrial functional assessments. These integral approaches combined with measurements of distinct components of the mitochondrial ATP-generating machinery have provided invaluable information on cardiac mitochondrial function under physiologic and pathophysiologic conditions.
MRS as a noninvasive in vivo approach complements in vitro studies of cardiac mitochondrial function. It has been used as a powerful research tool to assess myocardial energetics in different regions of the heart. Another emerging complementary noninvasive technology, which measures tissue oxygen consumption, is near-infrared spectroscopy [152–154]. Combining this method with 31P-MRS can assess mitochondrial coupling in vivo, which is currently technically challenging [155, 156]. In addition, it has been demonstrated that protein separation combined with MRS enables in vivo determination of synthesis rates of individual mitochondrial proteins [157]. However, transfer of MRS-based technologies into the clinical realm requires further improvement of reproducibility and reduction of acquisition times. Future technical, such as increased magnetic field strengths (>3-T), and data processing improvements will help to overcome the current limitations, and MRS will finally become a clinical reality.
The mitochondrial proteomics has emerged as one of the most active areas in mitochondrial studies today. Due to complimentary approaches, relied mainly on large-scale proteomics, high-resolution microscopy, and computational analysis, approximately 75% of mitochondrial proteins have been identified. High sensitivity of current MS techniques has highlighted the importance of efficient and reliable procedures for isolation of human mitochondria and especially of mitochondrial compartments, such as outer and inner membranes, cristae, matrix, and OXPHOS complexes. Future proteomic studies have to also address tissue-specific and developmental differences in the human mitochondrial proteome. Further careful proteomic analysis has to focus on various posttranslational modifications of mitochondrial proteins as an essential regulatory mechanism of mitochondrial function.
Another rapidly developing high-throughput technology is protein microarray. Based on the use of several thousands of proteins immobilized on a miniaturized solid surface, it enables sensitive, large-scale screening for profiling of protein expression, protein-protein interactions, posttranslational modifications, and specific cofactor requirements [158, 159]. This powerful technology needs to be applied for study mitochondrial proteome in the human heart.
In conclusion, the outlined technological approaches will help elucidate the complex nature of mitochondrial function, uncover the molecular basis of heterogeneous mitochondrial disorders associated with CVD, and define targets for their therapeutic treatment.
Summary
Cardiac mitochondria are complex highly organized cellular organelles, which play central roles not only in energy homeostasis but also in various biosynthetic, signaling, and cell death pathways. Moreover, mitochondria are highly dynamic organelles that continuously divide and fuse as well as move within the cell. Hence, a wide range of methodological approaches have been developed to assess mitochondrial functions.
Recent advances in fluorescent imaging technologies have significantly enhanced our ability to analyze mitochondrial morphology and dynamics and precisely measure levels of specific metabolites and ions within its sub-compartments. A variety of fluorescent probes and potentiometric dyes have been used to quantitatively evaluate overall cardiomyocyte mitochondrial number, membrane potential, oxidative stress, and Ca2+ concentrations.
While these fluorescent dye markers stain the whole organelle, a variety of FPs has been produced to analyze the dynamics of specific mitochondrial proteins in the living cells. Based on various FP variants, molecular sensors to measure number of mitochondrial parameters, such as redox potential, Ca2+ and Cl– levels, and pH, are currently available.
Recently, “nanoscopy” or “super-resolution” fluorescence technologies have been introduced to overcome the limiting role of diffraction in a lens-based optical microscopy and to provide nanometer-level precision coordinates. Several physical concepts relaying on reversible saturable optical fluorophore transitions have been developed, such as stimulated emission depletion microscopy and ground state depletion microscopy.
EM has tremendously advanced and during the last six decades become a powerful tool to study mitochondrial ultrastructure and function. Various modifications of the fixation, dehydration, sectioning, and staining of section better preserving the native mitochondrial morphology have been developed. Combination of EM with immunostaining techniques has significantly enhanced the power of this technique and has provided further insights into mitochondrial architecture and function.
Since conventional transmission EM generates 2D images of 3D objects, 3D imaging techniques such as high-resolution scanning EM and ET have been introduced. This revolutionizing approach is able to yield 3D reconstruction of mitochondria at molecular levels.
The newly emerging cryo-ET using quickly frozen samples is devoid of artifacts induced by chemical fixation, dehydration, and staining. Cryo-ET, combined with 3D image classification and single particle averaging, can visualize not only mitochondrial ultrastructure but also mitochondrial multiprotein complexes at near-atomic resolution.
Most of molecular and biochemical studies of cardiac mitochondria have relied on isolated organelles. In the past decades, numerous isolation and fractionation procedures of mitochondria as well as their membrane-bound sub-compartments together with the identification of specific markers have been reported. Isolated cardiac mitochondria retain their essential morphological and functional characteristics.< div class='tao-gold-member'>Only gold members can continue reading. Log In or Register to continue
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


