Databases in Cardiology
Michael S. Lauer
Eugene H. Blackstone
Overview
A clinical database is a systematic collection of observations, most usefully expressed as variable–value pairs, related to patients or clinical processes. With the advent of inexpensive, sophisticated computer technology, databases in cardiology can be used for case finding, information retrieval, description of cohorts, generation of new knowledge about heart disease and its treatment, and construction of predictive models for improving decision-making and informed consent for treatment.
Databases in cardiology have been developed in five major settings: (a) population-based studies, like the Framingham Heart Study, which are used to describe risk factors for, and natural history of, cardiovascular disease in the community; (b) disease-oriented registries, which describe care processes for, and outcomes of, patients with specific problems, like acute myocardial infarction; (c) procedure- or technology-oriented registries; (d) government-mandated and administrational databases; and (e) registries derived from randomized clinical trials. Newer database technologies, including object-oriented and semistructured databases, along with the computerized health record, represent means by which clinical database methods can be incorporated into routine clinical care.
Glossary
Case–control study
A study design in which patients with and without a specified outcome are identified and compared with respect to an exposure variable of interest.
Case-mix adjustment
Identifying and controlling for patient characteristics (case mix) and structural variables over which physicians and health care institutions have no control and that may explain variances in outcome.
Categorical variable
A variable that can have only a finite set of values.
Coding system
A system for describing outcomes, clinical findings, and therapies according to a widely accepted format, for example, Current Procedural Terminology (CPT), International Classification of Diseases, 9th revision (ICD-9), and Systematized Nomenclature of Medicine (SNOMED).
Cohort study
A study design in which a group of participants or patients is prospectively defined, divided according to the presence or absence of an exposure variable of interest, and followed over time for occurrence of prespecified outcomes. Generally, there are consecutive patients within a specified time frame with a given disease and treatment.
Collinearity
A phenomenon of multivariable models in which two or more variables that are essentially predictive of one another (correlation near 100%) lead to computational uncertainty of estimating model parameters; minor fluctuations of such variables may lead to major change in model parameter estimates.
Computerized patient record
A type of clinical database in which an electronic medium is used to capture, process, analyze, and retrieve values for variables representing the entire clinical process. In contrast to the electronic patient record, patient information is represented as values for variables.
Content validity
Appropriate and complete selection of covariates in a predictive model or of items in a quality-of-life instrument.
Continuous variable
A variable that can have an infinite set of values.
Cox proportional hazards model
A multivariable model that relates the logarithm of the unspecified hazard function ratio between groups of patients to a linear equation.
Data dictionary
A set of definitions for data elements.
Data element
A specific piece of organized information about a person or event.
Database design
A model according to which data elements are linked logically to one another.
Database standards
Agreed-on methods for defining terms and interfaces, thus allowing cross-communication between databases.
Face validity
The clinical sense of a prediction model, that is, the model makes sense, given current understanding.
Field
A placeholder for a specific value regarding a person or event in relational-type databases; synonymous with variable.
Flatfile
A single data table that contains columns representing variables and rows representing bases of observations, for example, individuals. The table contains no structural information about the data values themselves.
Hazard function
The instantaneous risk of a time-related event.
Hierarchical database
A database model based on a nested root-subordinate segment (tree) structure.
Interaction
A situation in which the strength of association between one variable and an outcome is affected by another variable; this is also known as effect modification or modulation. It is distinguished from confounding, in which an association between a variable and an outcome only reflects the association between another variable with the index variable and with outcome.
Internet
A worldwide system of computer networks that interact using a common communications protocol.
Logistic regression model
A multivariable model in which the logarithm of the odds of a time-fixed outcome event is related to a linear equation.
Metadata
Data about data, that is, information describing other data. In a database context, it is usually considered the structural information associated with both data and database organization.
Object-oriented database
A database model in which complex data structures (objects) are used that consist of both values for variables and instructions for operations to be performed on the data.
Ontology
A body of formally represented knowledge based on an abstract world view (e.g., National Library of Medicine Metathesaurus). In the context of databases, it distinguishes those that are based and searched on data contents versus those that are searched on data structure, such as metadata.
Outcomes management
A process by which clinical processes and outcomes are recorded systematically in large databases and then analyzed to detect variances in outcome that can be improved by altering those processes.
Overfitting
Using too many covariates for the number of outcome events in a multivariable model; this can lead to spurious associations.
Predictive modeling
A process by which a clinical database is used to describe mathematically the likelihood of outcome events, given a set of values for variables of a new patient.
Propensity analysis
A method of adjusting for possible confounding or selection biases using the probability of an exposure (or nonrandomized treatment) that is calculated based on numerous measurable nonexposure control variables.
Predictive validity
How well a multivariable model predicts outcome in a database separate from the one from which the model was derived.
RDF
Resource description framework, which describes metadata about specific nodes on a network (usually a Web site, but in a database context may be a semistructured data node).
Relational database
A database model that consists of multiple cross-referenced tables consisting of rows and columns that can be linked together by set(s) of user-defined “linking variables” and queried using Structured Query Language.
Reliability
Reproducibility or stability of data measures.
Semistructured data
Complex data elements whose contextual structures are embedded along with the values for variables, rather than being dependent on an external data structure. Data elements are simultaneously independent self-defining entities and self-organizing given a specific query.
Spreadsheet
A table of values arranged in rows and columns, in which values at row–column intersections (called cells) can have predefined relationships with other cells. Data structure accompanies the spreadsheet.
Validity
How well a data element or predictive model reflects what is supposed to be measured or predicted.
XML
Extensible Markup Language, a formal, concise, human-legible, machine (computer) and operating system–independent language that describes both data entities (objects) and the behavior of computer programs (XML Processor) that understand and present them. This is achieved through markup, which encodes (tags) a description of the entity’s storage layout and logical structure.
World Wide Web
A technology by which access to the Internet is made easy by use of graphical, hypertext-linked interfaces.
Introduction
In the late 1940s and early 1950s, epidemiologists from the U.S. Public Health Service and academic cardiologists and internists in the Boston area collaborated to develop a population-based database of cardiovascular disease in Framingham, Massachusetts (1,2). Early development of the Framingham Heart Study database was fraught with difficulties (3): Methods of population sampling were not well developed, the natural history of coronary artery disease (CAD) was poorly understood, clinical manifestations of CAD were known to only partially reflect pathologic events, the modern “computer era” had not yet begun, and modern biostatistical science was in its infancy. Nonetheless, the investigators successfully assembled a cohort of more than 5,000 adults who have now been systematically followed for nearly 50 years (1); later, in the 1970s, a new cohort based on the offspring and spouses of offspring of the original cohort was assembled, and it continues to be followed (4).
The Framingham Heart Study illustrates many attributes of successful databases (5): The database was assembled and maintained by a large, multidisciplinary team with involvement of clinicians, epidemiologists, computer scientists, and biostatisticians. Funding was adequate and stable, and goals were well defined, leading to high-quality, focused data collection. Efforts have been made to keep the database relevant with appropriate addition, alteration, and deletion of data fields as new technologies and concepts change the questions that investigators wish to answer. Perhaps most important, dynamic and strong leaders have championed the development, implementation, use, and analyses of the database and have publicly advocated its benefits.
With the advent of the computer, large databases became possible in clinical environments as well. Today, clinical databases have expanded into multiple areas of practice and investigation, presenting tremendous opportunities for improving knowledge about and care of patients with cardiovascular disease, but also frustrating developers and users with numerous technical and design challenges.
What is a Database? Database Technology, Terms, and Definitions
A clinical database is a systematic collection of facts about individual patients or clinical events (Table 44.1). Computer-based databases divide these facts into (a) what the fact is, which can be termed a variable or, in specific contexts, a field or column name; (b) the specific value for the variable for an individual patient; and (c) structural information, including the name of the data element, unique identifier for the data element, version, authorization information, language, definition, obligation (whether the data element is required or not), data type (e.g., numerical or character), limits (e.g., age cannot be <0 or >120), author (name of who or what obtained and recorded the information), time-stamp, temporal attributes, and many other attributes of the variable and value. A collection of data elements that relate to one specific person or clinical event constitutes a patient record.
Types of Database Elements
Clinical database variables can be either categorical (nominal) or continuous. Categorical variables may be limited to only two possible values and thus be dichotomous; examples include yes or no, 0 or 1, male or female, and presence or absence of chronic heart failure. Alternatively, categorical variables may be ordinal, that is, have a limited list of possible values that bear an ordered relation to one another; examples include New York Heart Association functional class and severity of mitral valve regurgitation (6). Finally, categorical variables may be polytomous, that is, have an unordered but finite list of possible values, such as etiology of subacute bacterial endocarditis or names of bones in the body. Continuous variables can hold an infinite number of values; examples are age and systolic blood pressure.
TABLE 44.1 Basic Concepts of Database Construction | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||||||||||
Data Dictionaries and Metadata
If data are to be understood, valid, and reliable, it is essential that all data elements be defined accurately. A common means of accomplishing this is to create a separate computer file that contains the list of all files in the database, a list of all variables, definitions and data type of each variable, and lists and definitions of all possible values with each variable. Such a file is known as a data dictionary (7).
With the advent of worldwide exchange of information, attention has focused on data definitions and formatting that are independent of specific machine types and operating systems using internationally agreed on standards. Data definitions are now more fully expressed in terms of metadata. Metadata are “data about data” that contain all structural attributes of the data and database. Metadata may be separated from the data to form part of the structure of the database, in which case data elements are simple, consisting of just the value, or metadata may be directly associated with each data element, in which case data elements may be complex (semistructured data). Metadata may be further organized with interrelationships to become a knowledge base or ontology.
Structure and Content of Clinical Databases
Determination of the size, structure, and content of clinical databases has been a major source of frustration, in part because no standards for these existed before the modern computer age. Ellwood described certain rules that should be followed if a large clinical database is to remain robust over time (8). The goals of assembling the database have to be prospectively determined to define appropriate data elements. The size of the database must be carefully considered; large databases contain more information, but excessively large numbers of data elements can lead to significant deterioration of data quality. Although data may be collected in many geographic locales, a mechanism for centralizing data is essential. Finally, the ideal clinical database is not confined to hospital events. It is the frustration with the predefinition of databases, which presupposes the relations and types of questions likely to be asked of it in the future, that has stimulated development of infinitely extensible databases, such as exemplified by the World Wide Web and semistructured databases.
Database Standards
For clinical data obtained outside the confines of a clinical trial or registry to be considered reliable for analyses, it is essential for standards to be developed to allow data fields to be consistent between different databases and to allow data to be transferred between systems (9). Numerous coding systems (such as ICD-9, SNOMED, Unified Medical Language System [UMLS], CPT, and others) for clinical databases have been developed to represent clinical classifications and nomenclatures (10,11).
Database Designs
A database “design” refers to the model according to which data fields are logically linked to one another (12); the most commonly cited database designs are flatfile, spreadsheet, hierarchical, relational, and object oriented. The simplest design is a single table of values, called a flatfile. A flatfile is non–self-documenting, requiring a separate data dictionary to accompany the data file. The next-simplest design is the spreadsheet. It is superior to the flatfile not only in containing self-documentation, such as column headings, but also in allowing in-place associations to be established among the data cells. A major limitation of flatfiles and spreadsheets is that they have primarily been proprietary products, and they allow entry of data values in an unstructured fashion that may permit human understanding of the data but not computer understanding necessary for analysis. Another early design was the hierarchical model, which consisted in linking multiple collections of similar information into a tree structure, with root and child nodes as branches and ending with leaf nodes. A major disadvantage of this database design is that hierarchies are arbitrary and limiting.
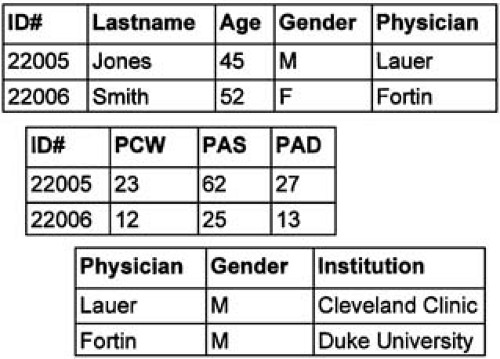
A database model that ties together multiple tables of information and provides more flexibility is the relational model. Relational databases allow for efficient, powerful storage of large quantities of information (13,14). Tables within a relational database are linked by user-defined cross-references or “linking variables” (Fig. 44.1). Relational databases are, however, static repositories. Imagine a database that could note that an entered value for a glucose tolerance test was well above the normal range and therefore set into motion a predetermined sequence of activities, such as paging the physician, printing a diabetic diet based on patient characteristics, discovering a clinical trial of pancreatic islet transplantation, or a host of other functions. This is possible in object-oriented databases. Objects contain not only data, but also instructions on how to react to certain data values.
Although the relational model is the most pervasive in clinical databases today, two other challengers deserve mention. One is an old technology, time-oriented databases, and the other is new, semistructured databases. Semistructured databases permit the self-assembly of an ad hoc structured database at the time of an analysis using information embedded in the data elements themselves. This is the exact opposite of the traditional approach to structured databases, in which the database is fixed and, after some lapse of time, queried and an analysis performed.
Data Quality
Missing Data
Rigorously run prospective, randomized clinical trials and some multicenter registries have specific mechanisms in place to ensure comprehensive data collection. Observational clinical databases in which data are obtained as part of clinical care are often plagued, however, by missing data, which can seriously bias accurate analyses and development of prediction models (Table 44.2) (8,15,16).
As described by Little and Rubin (17), missing data can be classified into three specific types: (a) missing completely at random, in which patients with missing data cannot be distinguished in any way from those without missing data; (b) missing at random, in which the pattern of missing data is due to some external cause unrelated to the variable with missing data; and (c) not missing at random, in which the mechanism by which data are missing is directly linked to the variable itself. An example of the last case is lack of stress-testing data in a patient with cardiogenic shock. A number of methodologies have been developed for managing missing data, ranging from simple imputation, nearest-neighbor matching, propensity analysis, informed imputation based on regression modeling, use of dummy variables to denote missingness, and multiple imputation.
Data Credibility
When considering accuracy of data, one must assess both reliability and validity. Reliability refers to how stable or
reproducible measurements are. If several independent reviewers code a certain set of measurements or abstract information from a medical record in similar ways with the same result, the data can be considered reliable. Validity, on the other hand, refers to whether the data represent what they are supposed to measure. For example, in one database the diagnosis of “myocardial infarction” was incorrect in 57% of recorded cases (18).
reproducible measurements are. If several independent reviewers code a certain set of measurements or abstract information from a medical record in similar ways with the same result, the data can be considered reliable. Validity, on the other hand, refers to whether the data represent what they are supposed to measure. For example, in one database the diagnosis of “myocardial infarction” was incorrect in 57% of recorded cases (18).
TABLE 44.2 Limitations of Observational Data | ||||||||
|---|---|---|---|---|---|---|---|---|
|
What are Clinical Databases Used for?
Fundamental Uses of Clinical Databases
As described by Safran (19), there are four fundamental uses of clinical databases: (a) results reporting, (b) case finding, (c) cohort description, and (d) predictive modeling. An additional use that has gained much recent attention is for assessment of medical care quality (20). Results reporting refers to the ability to retrieve information about a specific patient or procedure; this is the type of activity by which practicing clinicians would most often interact with, or at least “seek rewards” from, a clinical data repository. Case finding involves looking for patients with similar problems or outcomes as a patient of interest, often to direct management based on prior individual experiences. Cohort description seeks to simply describe a group of patients or procedures based on a common set of attributes. Prediction modeling involves applying sophisticated analytical methods to describe changes or associations that can lend insight into the natural history of disease processes, the effects of different clinical interventions, and/or quality of care within given institutions or by specific providers. It is this use of databases that presents the most powerful, and perhaps also the most controversial, benefit of computerized clinical information technology.
Outcomes Management
In the 1988 Shattuck Lecture to the Annual Meeting of the Massachusetts Medical Society, Paul Ellwood advocated the nationwide adoption of outcomes management, which he described as a new “technology of patient experience” (8). Work by Wennberg and others (21,22,23) that showed tremendous variation of practice patterns without clear differences in patient outcome has caused the public and third-party payers to demand better-quality clinical effectiveness information. Physicians, too, are frustrated by their inability to accurately and quantitatively predict the effect of their care on patient outcomes because good-quality data from randomized, controlled trials simply do not exist for many common clinical scenarios (24). To help to solve these problems, the process of outcomes management would seek to use high-quality data based on real patient experiences to help physicians, payers, and patients make the most rational treatment choices.
Why Not Just Use Randomized, Controlled Trials to Determine Clinical Effectiveness?
A major component of outcomes management is the determination, based on analyses of large observational clinical databases, of how outcomes are associated with physician plan and hospital execution factors after “adjusting for” patient factors. Some have argued, however, that using observational databases to assess the effect of therapies or behaviors under physician or patient control is inherently flawed and that only randomized trials can appropriately answer these questions (25). The main strengths of randomized, controlled trials include (a) assembly of a single, well-defined inception cohort; (b) elimination of potential biases and confounding, particularly those caused by unknown or unmeasurable factors; and (c) adherence to well-defined clinical care and data collection protocols with vigorous quality assurance and safety mechanisms in place (26). However, as reviewed by Califf and colleagues (27), clinical trials suffer from several inherent limitations: (a) they are expensive and sometimes very time consuming; (b) therapies change such that by the time trial results are reported, new technologies may have become available that may replace the methods tested, (c) many patients, particularly those with comorbidities or the elderly, are excluded, (d) trials are performed more often in patients at tertiary referral centers, raising generalizability questions, and (e) the benefits of a therapy as noted in the ideal conditions of a clinical trial may not necessarily apply to the “real world” of clinical care. Furthermore, some clinical treatment questions simply cannot be answered with randomized trials. For example, a recent study found that a prolonged door-to-balloon time was associated with higher mortality among patients undergoing primary angioplasty for acute myocardial infarction (28). It is hard to imagine that the validity of this finding could be tested by a controlled trial in which patients would be randomized to an immediate versus delayed angioplasty or even to a series of prescribed intervals (24).
Causes of Bias
Although some have found that analyses of clinical databases can lead to conclusions that are consistent with those from presumably unbiased, randomized clinical trials (29,30,31), the presence of bias remains a major impediment to the widespread acceptance of clinical databases as a valid source of insights on clinical effectiveness (25,26,32




Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree