Apolipoprotein E, a protein component of blood lipid particles, plays an important role in lipid transport. Different mutations in the apolipoprotein E gene have been associated with various clinical phenotypes. In an initiated study of Qataris, we observed that 17% of the African-derived genetic subgroup were heterozygotes for a rare Arg145Cys (R145C) variant that functions as a dominant trait with incomplete penetrance associated with type III hyperlipoproteinemia. On the basis of this observation, we hypothesized that the R145C polymorphism might be common in African-derived populations. The prevalence of the R145C variant was assessed worldwide in the “1000 Genomes Project” and in 1,012 whites and 1,226 African-Americans in New York, New York. The 1000 Genomes Project data demonstrated that the R145C polymorphism is rare in non–African-derived populations but present in 5% to 12% of Sub-Saharan African-derived populations. The R145C polymorphism was also rare in New York whites (1 of 1,012, 0.1%); however, strikingly, 53 of the 1,226 New York African-Americans (4.3%) were R145C heterozygotes. The lipid profiles of the Qatari and New York R145C heterozygotes were compared with those of controls. The Qatari R145C subjects had higher triglyceride levels than the Qatari controls (p <0.007) and the New York African-American R145C subjects had an average of 52% greater fasting triglyceride levels than the New York African-American controls (p <0.002). From these observations, likely millions of people worldwide derived from Sub-Saharan Africans are apolipoprotein E R145C. In conclusion, although larger epidemiologic studies are necessary to determine the long-term consequences of this polymorphism, the available evidence suggests it is a common cause of a mild triglyceride dyslipidemia.
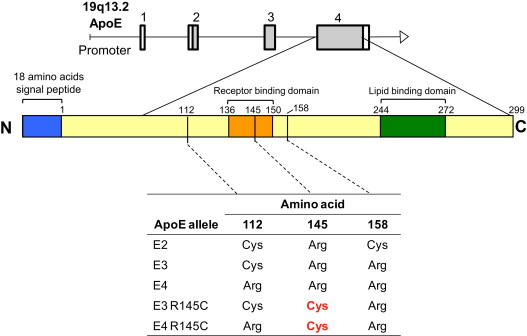
Apolipoprotein E (ApoE), a major component of blood chylomicrons, very-low-density lipoprotein, and high-density lipoprotein (HDL) particles, has several alleles that differ by point mutations that are associated with a range of clinical phenotypes ( Supplemental Table 1 ). As part of a study to assess the genetic variations of medical importance among Qataris, a population that evolved at the migration crossroads of Africa and characterized by a high prevalence of type 2 diabetes mellitus, obesity, and cardiovascular disease, we observed a high frequency of a point mutation in amino acid 145 of ApoE, in which arginine is substituted by cysteine (R145C). Since its discovery in 1982, the mutation has been described in 32 subjects ( Supplemental Table 2 ) as inherited in a dominant fashion with incomplete penetrance and hypothesized to be a rare cause of type III hyperlipoproteinemia. The Qatari population is composed of 3 distinct genetic subpopulations, including Arab, Persian, and African (Q3). Recognizing that all the R145C variants were in Qataris of African genetic ancestry, we hypothesized that the R145C polymorphism might be far more common in those of African descent than previously recognized. To assess this, we evaluated the genomes represented by the “1000 Genomes Project” and of 1,012 whites and 1,226 African-Americans in the New York City Metropolitan Area. The prevalence of the mutation in the study population suggests that ApoE R145C might put large numbers of Africans and African descendants worldwide at risk of a mild triglyceride dyslipidemia.
Methods
Under protocols approved by the Weill Cornell New York and Weill Cornell-Qatar institutional review boards and the Hamad Medical Corporation Ethics Committee (Doha, Qatar), all participants in Qatar and New York signed informed consent documents for participation in the present study. Three populations were assessed: Qataris (n = 228; subgrouped into 3 distinct genetic subpopulations: Arabian, n = 123; Persian, n = 82; and African [Q3], n = 23), worldwide subjects from the 1000 Genomes Project (n = 1,094), and whites (n = 1,012) and African-Americans (n = 1,226) in the New York metropolitan area ( Supplemental Methods ).
ApoE variants were identified in the Qatari and New York African-American populations using polymerase chain reaction ( Supplemental Table 3 ) and sequencing, when necessary. The ApoE variants in the 1000 Genomes Project data were identified from the single nucleotide polymorphisms and, when available, sequences ( Supplemental Methods ).
The clinical phenotypes of the Qatari Q3 R145C were identified from a review of medical records, including 4 to 14 fasting lipid profiles within 7 to 9 years (i.e., triglycerides, cholesterol, low-density lipoprotein [LDL] cholesterol, HDL cholesterol; Supplemental Table 4 ). The Q3 R145C subjects were compared with 15 Q3 controls for whom lipid levels were available (1 to 10 fasting lipid profiles within 1 to 9 years).
Of the 53 New York African-American R145C, 23 had fasting routine lipid values available from the medical records (73% with 1 measurement and 27% with 2 to 47 measurements within 2 to 12 years). Of the African-American controls, 72 randomly chosen subjects had fasting lipid studies retrieved from their medical records (71% had 1 study and 29% had 2 to 36 studies within 1 to 12 years). Attempts were made to interview all 53 New York African-American R145C subjects. Of these 53, 16 (30%) returned for additional studies. Of these 16, 5 also had previous values available from their medical records. The fasting plasma from these 16 R145C subjects was assessed for lipoprotein subclass profiling, hemoglobin A1C, high-sensitivity C-reactive protein, lipoprotein (a), ApoA1, ApoB, thyroid-stimulating hormone, and glucose using proton nuclear magnetic resonance spectroscopy (Liposceince, Raleigh, North Carolina), and for LDL electrophoresis and subfractionation (Berkeley HeartLab, Alameda, Calif).
The fasting lipid studies of the R145C subjects and controls were compared using 2 methods: (1) using all mean values from each subject, with a single-factor analysis of variance (ANOVA) and (2) weighting the mean value for each subject by the variance in mean response, with a nested ANOVA ( Supplemental Methods ).
Results
ApoE2, ApoE3, and ApoE4 represent >95% of ApoE alleles worldwide. In most populations, ApoE3 is the most common, followed by ApoE4 and then ApoE2 ( Figure 1 ). Of the common ApoE alleles in the overall Qatari population, ApoE3 dominated, with ApoE4 more common than ApoE2 ( Table 1 ). However, in the context of the Qatari genetic subpopulations, the proportion of the common ApoE alleles differed (p <10 −3 , chi-square), with the greatest percentage of the ApoE3 allele in the Persian group (93%), the greatest percentage of the ApoE2 allele in the Q3 (African) group (11%), and the greatest percentage of the ApoE4 allele also in the Q3 group (15%).
| ApoE Genotypes and Alleles ∗ | All Qataris | Q1 (Arab) | Q2 (Persian) | Q3 (African) |
|---|---|---|---|---|
| Genotype | ||||
| Total | 228 (100) | 123 (100) | 82 (100) | 23 (100) |
| E2/E2 | 1 (0.4) | 1 (0.8) | 0 (0) | 0 (0) |
| E3/E3 | 169 (74.1) | 91 (74.0) | 70 (85.3) | 8 (34.8) |
| E4/E4 | 1 (0.4) | 1 (0.8) | 0 (0) | 0 (0) |
| E2/E3 | 13 (5.7) | 3 (2.4) | 6 (7.3) | 4 (17.4) |
| E2/E4 | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| E3/E4 | 38 (16.7) | 26 (21.1) | 6 (7.3) | 6 (26.1) |
| E3/E5 † | 1 (0.4) | 1 (0.8) | 0 (0) | 0 (0) |
| E2R150H/E2 ‡ | 1 (0.4) | 0 (0) | 0 (0) | 1 (4.3) |
| E3R145C/E3 § | 3 (1.3) | 0 (0) | 0 (0) | 3 (13.0) |
| E3R145C/E4 ‖ | 1 (0.4) | 0 (0) | 0 (0) | 1 (4.3) |
| All R145C heterozygotes | 4 (1.8) | 0 (0) | 0 (0) | 4 (17.4) |
| Alleles | ||||
| Total | 456 (100) | 246 (100) | 164 (100) | 46 (100) |
| E2 | 16 (3.5) | 5 (2.0) | 6 (3.7) | 5 (10.9) |
| E3 | 393 (86.2) | 212 (86.2) | 152 (92.7) | 29 (63.0) |
| E4 | 41 (9.0) | 28 (11.4) | 6 (3.7) | 7 (15.2) |
| E5 | 1 (0.2) | 1 (0.4) | 0 (0) | 0 (0) |
| E2R150H | 1 (0.2) | 0 (0) | 0 (0) | 1 (2.2) |
| E3R145C | 4 (0.9) | 0 (0) | 0 (0) | 4 (8.7) |
| All R145C heterozygotes | 4 (0.9) | 0 (0) | 0 (0) | 4 (8.7) |
∗ The DNA for all Qataris was first assessed using TaqMan polymerase chain reaction at single nucleotide polymorphisms rs429358 (amino acid 112), rs769455 (amino acid 145), and rs7412 (amino acid 158). This was followed by sequencing of all ApoE exons and flanking introns. If heterozygosity was present at 2 single nucleotide polymorphisms in the same subject, cloning and complete sequencing was performed to unambiguously identify the genotype.
† The E5 allele (E212K on the E3 background) has been previously described ; this subject had no dyslipidemia.
‡ The E2R150H (R150H substitution on the E2 background) allele was novel to the present study; this subject had no dyslipidemia.
§ See Supplemental Table 4 for demographics and values for each subject of the 3 E3R145C/E3 heterozygotes (age 52.7 ± 2.8, 1 man and 2 women, body mass index 35.3 ± 1.6 kg/m 2 ).
‖ For the E3R145C/E4 heterozygote, 1 subject was a 40-year-old woman, with a body mass index of 48.6 kg/m 2 ; see Supplemental Table 4 for additional details.
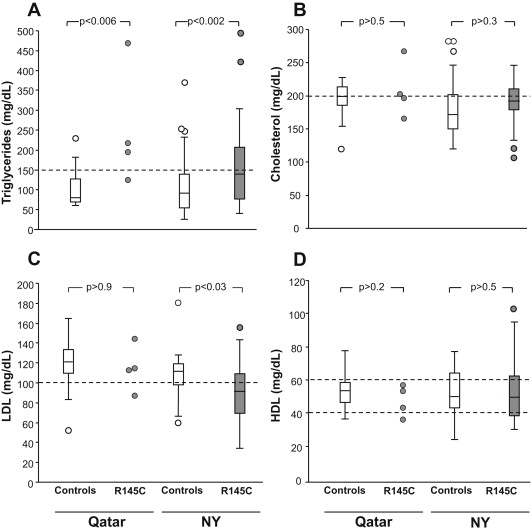
Of the 456 Qatari ApoE alleles assessed, 4 (0.9%) had the R145C polymorphism ( Table 1 ). All were on the ApoE3 background, all were heterozygotes, and all were in the Q3 (African) subpopulation, representing 17.4% of all Q3 subjects. All 4 subjects had type 2 diabetes, all were obese, 1 had cataracts, 2 had cardiovascular disease, and 1 had kidney disease ( Supplemental Table 4 ). None had thyroid disease or xanthomas; 3 had hypertriglyceridemia (mean level 193 to 465 mg/dl), 2 of the 4 had hypercholesterolemia, 1 had low HDL cholesterol, and 3 of the 4 had elevated LDL cholesterol. Of the 15 Q3 Qatari controls, 4 (27%) had type 2 diabetes compared with 100% of the Qatari R145C subjects (p <0.04). The incidence of xanthomas, cataracts, hypertension, and thyroid disease was similar in the control group and R145C group (p >0.1, for all comparisons). The Qatari R145C triglyceride levels were significantly greater than those in the Qatari controls (p <0.006, single-factor ANOVA, p <0.007 nested ANOVA). The cholesterol, HDL, and LDL levels were not significantly different between the 2 groups (p >0.2, all comparisons, single-factor or nested ANOVA; Figure 2 ).
Assessment of the 1000 Genomes Project populations revealed that 12 of the 97 (12%) Luhya subjects in the Webuye, Kenya population and 7 of the 88 (8%) Yoruban subjects in Ibadan, Nigeria were R145C ( Table 2 ). In addition, 3 of 61 (5%) of African ancestry in the Southwest United States population were R145C. The R145C polymorphism was not observed in the British, Finnish, Northern and Western European ancestry, Iberian populations in Spain, Toscani in Italy, Han Chinese in Beijing, China, Han Chinese South, Japanese in Tokyo, Japan, or Colombian in Medellin, Colombia, populations and was only rarely present in the Puerto Rican and Mexican populations (1.5% to 1.8%).
| Variable | Europe † | Asia ‡ | America § | Africa ‖ | All 1000 Genome Project Subjects | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CEU | FIN | GBR | IBS | TSI | CHB | CHS | JPT | ASW | CLM | MXL | PUR | LWK | YRI | ||
| Genotype | |||||||||||||||
| Total | 87 (100) | 93 (100) | 89 (100) | 14 (100) | 98 (100) | 97 (100) | 100 (100) | 89 (100) | 61 (100) | 60 (100) | 66 (100) | 55 (100) | 97 (100) | 88 (100) | 1,094 (100) |
| E2/E2 | 0 | 0 | 1 (1.1) | 0 | 0 | 0 | 0 | 0 | 1 (1.6) | 1 (1.7) | 0 | 0 | 1 (1.0) | 0 | 4 (0.4) |
| E2/E3 | 10 (11.5) | 13 (14.0) | 10 (11.2) | 0 | 8 (8.2) | 18 (18.6) | 16 (16.0) | 8 (9.0) | 10 (16.4) | 6 (10.0) | 8 (12.1) | 7 (12.7) | 8 (8.2) | 15 (17.0) | 137 (12.4) |
| E2/E4 | 1 (1.1) | 0 | 2 (2.2) | 1 (7.1) | 1 (1.0) | 2 (2.1) | 1 (1.0) | 0 | 4 (6.6) | 1 (1.7) | 0 | 0 | 6 (6.2) | 9 (10.2) | 28 (2.7) |
| E3/E3 | 51 (58.6) | 50 (53.8) | 54 (60.7) | 9 (64.3) | 72 (73.5) | 57 (58.8) | 66 (66.0) | 67 (75.3) | 28 (45.9) | 35 (58.3) | 46 (69.7) | 36 (65.5) | 27 (27.8) | 35 (39.8) | 633 (57.8) |
| E3/E4 | 24 (27.6) | 27 (29.0) | 19 (21.3) | 4 (28.6) | 17 (17.3) | 20 (20.6) | 17 (17.0) | 12 (13.5) | 13 (21.3) | 16 (26.7) | 10 (15.2) | 11 (20.0) | 31 (32.0) | 21 (23.9) | 242 (22.2) |
| E4/E4 | 1 (1.1) | 3 (3.2) | 3 (3.4) | 0 | 0 | 0 | 0 | 2 (2.2) | 2 (3.3) | 1 (1.7) | 0 | 0 | 5 (5.2) | 1 (1.1) | 18 (1.6) |
| E3/E3R145C | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 (4.9) | 0 | 1 (1.5) | 1 (1.8) | 2 (2.1) | 4 (4.5) | 11 (1.0) |
| E4/E3R145C | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 (4.1) | 2 (2.3) | 6 (0.5) |
| E3/E4R145C | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 (2.1) | 0 | 2 (0.2) |
| E3/E4R158C | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 (1.5) | 0 | 2 (2.1) | 0 | 3 (0.3) |
| E3R145C/E4R145C | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 (1.0) | 0 | 1 (0.1) |
| E4/E4R145C | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 (1.0) | 1 (1.1) | 2 (0.2) |
| E4/E4R145CR158C | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 (1.0) | 0 | 1 (0.1) |
| E4/E4R158C | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 (5.2) | 0 | 5 (0.5) |
| E4R145C/E4R158C | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 (1.0) | 0 | 1 (0.1) |
| Total R145C genotypes | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 (4.9) | 0 | 1 (1.5) | 1 (1.8) | 12 (12.4) | 7 (8.0) | 24 (2.2) |
| Alleles | |||||||||||||||
| Total | 174 (100) | 186 (100) | 178 (100) | 28 (100) | 196 (100) | 194 (100) | 200 (100) | 178 (100) | 122 (100) | 120 (100) | 132 (100) | 110 (100) | 194 (100) | 176 (100) | 2,188 (100) |
| E2 | 11 (6.3) | 13 (7.0) | 14 (7.9) | 1 (3.6) | 9 (4.6) | 20 (10.3) | 17 (8.5) | 8 (4.5) | 16 (13.1) | 9 (7.5) | 8 (6.1) | 7 (6.4) | 16 (8.2) | 24 (13.6) | 173 (7.9) |
| E3 | 136 (78.2) | 140 (75.3) | 137 (77.0) | 22 (78.6) | 169 (86.2) | 152 (78.4) | 165 (82.5) | 154 (86.5) | 82 (67.2) | 92 (76.7) | 112 (84.8) | 91 (82.7) | 99 (51.0) | 110 (62.5) | 1,661 (75.9) |
| E4 | 27 (15.5) | 33 (17.7) | 27 (15.2) | 5 (17.9) | 18 (9.2) | 22 (11.3) | 18 (9.0) | 16 (9.0) | 21 (17.2) | 19 (15.8) | 10 (7.6) | 11 (10.0) | 58 (29.9) | 35 (19.9) | 320 (14.6) |
| E3R145C | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 (2.5) | 0 | 1 (0.8) | 1 (0.9) | 7 (3.6) | 6 (3.4) | 18 (0.8) |
| E4R145C | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 (2.6) | 1 (0.6) | 6 (0.3) |
| E4R145C.R158C ¶ | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 (0.5) | 0 | 1 (0.05) |
| E4R158C # | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 (0.8) | 0 | 8 (4.1) | 0 | 9 (0.4) |
| Total R145 alleles | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 (2.5) | 0 | 1 (0.8) | 1 (0.9) | 13 (6.7) | 7 (4.0) | 25 (1.1) |
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


