Single nucleotide polymorphisms (SNPs) at chromosome 4q25 (near PITX2 ) are strongly associated with atrial fibrillation (AF). We assessed whether a 4q25-tagging SNP (rs2200733) is associated with PR interval duration in patients with lone and typical AF and controls. Patients with lone (n = 169) and typical (n = 269) AF enrolled in the Vanderbilt AF registry and controls (n = 1,403) derived from the Vanderbilt DNA Biobank were studied. Carriage of the rs2200733T allele (CT or TT genotype) was more common in patients with lone (39%) than typical (25%) AF or controls (21%, p <0.01 for both comparisons). In both AF cohorts, we observed an association between genotype and PR interval duration (median PR interval for CC, CT, and TT: 162, 178, and 176 ms, respectively, for lone, p = 0.038 and 166, 180, and 196 ms, respectively, for typical, p = 0.001). After adjustment for covariates, the association between T allele and PR prolongation persisted, with mean effect size of 10.9, 12.8, and 4.4 ms for patients with lone and typical AF and controls, respectively (p <0.05 for each comparison). We found that a common 4q25 AF susceptibility allele (rs2200733) is associated with PR interval prolongation in patients with lone and typical AF and controls with no AF. Given that prolonged PR interval is an established risk factor for AF, this observation, in the context of previously described functional effects of PITX2 deficiency, provides further knowledge about the pathophysiological link of 4q25 variants with AF.
Atrial fibrillation (AF) is becoming increasingly common, with prevalence in the United States projected to roughly double by the year 2050 to an estimated 6 to 12 million. Identifying patients at high risk for developing AF, with the objective of developing strategies for prevention, is an important area of discovery. Genome-wide association studies have identified novel variants associated with prevalent AF. The strongest and most consistently associated locus is at chromosome 4q25 near the paired-like homeodomain transcription factor 2 ( PITX2 ) gene. Prolongation of the electrocardiographic PR interval has emerged as an important independent risk factor for AF and, in some patients, might represent an “endophenotype” or a heritable early marker of AF. Recently, a 4q25 variant was associated with PR-interval prolongation in patients with lone AF. Here, we sought to investigate the role of the rs2200733 variant at 4q25 on PR duration in 3 patient cohorts: those with lone AF, patients with typical AF, and controls free from AF or structural heart disease.
Methods
Patients with lone AF, typical AF, and controls with no AF were studied. Lone AF was defined as AF diagnosed before the age of 66 years in the absence of hypertension, hyperthyroidism, structural heart disease, or other predisposing cardiopulmonary disorders. Typical AF was characterized by age at diagnosis ≥66 years or the presence of hypertension, structural heart disease, or other predisposing cardiopulmonary disorders. Patients with lone and typical AF were prospectively enrolled in the Vanderbilt AF Registry, and all data were entered prospectively by dedicated study nurses. Controls without AF were identified by querying the Vanderbilt University Medical Center DNA Biobank (BioVU). This resource consists of de-identified medical records of Vanderbilt inpatients and outpatients and DNA extracted from blood that is left over after routine laboratory testing and scheduled to be discarded. As of August 2013, BioVU included >158,000 patients with available DNA. Controls were identified in BioVU using a previously validated algorithm that includes natural language processing, billing codes, and medication records. This automated algorithm was previously created and optimized through multiple reiterations with a manual review of medical records until a positive predictive value for AF of >95% was achieved. Patients with AF and controls identified by the automated algorithm were previously studied to confirm known genotype-phenotype relations. The Vanderbilt Institutional Review Board approved this study, and all participants gave written informed consent.
Clinical co-morbidities, including diabetes, hypertension, heart failure, coronary artery disease, myocardial infarction, and tobacco use were prospectively entered into the AF registry using standard definitions. Use of medications was entered on enrollment and updated periodically. In the no-AF cohort, information about co-morbidities and medications was queried using previously validated computer algorithms using natural language processing, laboratory values, medication records, and International Classification of Diseases–9 codes.
PR-interval duration, the study’s primary end point, was derived from 12-lead electrocardiograms (ECGs), where it was calculated using standard automated interpretation algorithms. For patients with lone and no AF, PR interval was ascertained from the ECG recorded at the time the patients were enrolled into the AF registry, unless the patient was not in sinus rhythm during this recording, in which case the first preceding ECG in which the patient was in sinus rhythm was used. In the lone AF cohort, medication records were manually reviewed to confirm that the index ECG was recorded while the patient was not currently or recently (≤7 days) on any antiarrhythmic drugs or atrioventricular nodal blockers. For patients with typical AF, medical records were manually searched to select ECGs recorded before the diagnosis of AF and while the patient was not on antiarrhythmic drug therapy. As many of the patients with typical AF had structural heart disease, it was not possible to exclude patients taking β blockers. Patients with lone or no AF who were taking β blockers were excluded from the study.
Genotyping was performed for the rs2200733 SNP, which was previously demonstrated to be an effective tagging SNP for the AF-associated 4q25 haplotype. In the AF registry, genotyping was performed using Taqman chemistry (Applied Biosystems, Foster City, California). Genotyping in the no-AF BioVU cohort was performed using the Human660W-Quad microarray platform (Illumina, San Diego, California). Laboratory personnel performing genotyping were blinded to clinical and electrocardiographic data.
Group comparisons were made using the Kruskal-Wallis test or Pearson correlation, as appropriate, for continuous variables or chi-square test for categorical variables. Multivariate linear regression was used to adjust for covariates. In univariate analysis, the following variables were associated with PR interval for each group and included in the final regression model: age and β-blocker use for typical AF and age, gender, and body mass index for no AF. In the smaller lone AF group, no clinical variable was associated with PR interval in univariate analysis. For consistency, age, gender, and body mass index were included in the multivariate model for lone AF. Nominal statistical significance was taken as a 2-tailed p ≤0.05. Analysis was performed using SPSS (v20, IBM Corp., Armonk, New York).
Results
Baseline demographic and clinical characteristics are summarized in Table 1 . Left atrial diameter was significantly greater in patients with lone AF homozygous for the rs2200733T allele compared with other genotypes. Otherwise, there were no statistically significant differences in baseline characteristics by genotype. Genotype frequencies for rs2200733 (CC, CT, and TT) were 0.61, 0.33, and 0.05 in lone AF, 0.75, 0.24, and 0.02 in typical AF, and 0.79, 0.20, and 0.01 in controls and were in Hardy-Weinberg equilibrium. Carriage of the T allele (genotypes CT and TT) was associated with lone (p <0.01) but not typical AF.
| Characteristic | Total | CC | CT | TT | p |
|---|---|---|---|---|---|
| Lone AF | n = 169 | n = 103 | n = 58 | n = 8 | |
| Age (yrs) | 54.5 ± 13.3 | 55.4 ± 13 | 54 ± 12.6 | 62 ± 7 | 0.53 |
| Men (%) | 115 (68) | 73 (71) | 36 (62) | 6 | 0.58 |
| Body mass index (kg/m 2 ) | 28.9 ± 6.2 | 28.2 ± 6.3 | 30.2 ± 6.3 | 32.1 ± 4.1 | 0.24 |
| Ejection fraction (%) | 57 ± 7 | 57 ± 7 | 57 ± 7 | 55 ± 5 | 0.62 |
| Left atrial diameter (mm) | 39 ± 7 | 39 ± 6 | 38 ± 8 | 46 ± 2 | 0.03 |
| Typical AF | n = 269 | n = 201 | n = 64 | n = 4 | |
| Age (yrs) | 66.6 ± 10.5 | 66.6 ± 11 | 66.4 ± 9.6 | 74 ± 3.2 | 0.30 |
| Men (%) | 207 (77) | 158 (79) | 46 (72) | 3 | 0.54 |
| Body mass index (kg/m 2 ) | 28.3 ± 7 | 28.6 ± 5.4 | 28.9 ± 7.5 | 30.6 ± 8.3 | 0.98 |
| Ejection fraction (%) | 43 ± 15 | 42 ± 15 | 46 ± 14 | 50 ± 14 | 0.07 |
| Left atrial diameter (mm) | 45 ± 8 | 45 ± 9 | 44 ± 6 | 44 | 0.93 |
| Heart failure (%) | 70 (26) | 55 (27) | 15 (23) | 0 | 0.43 |
| Diabetes (%) | 99 (37) | 78 (39) | 19 (30) | 2 | 0.40 |
| Coronary artery disease (%) | 145 (54) | 110 (55) | 34 (53) | 1 | 0.49 |
| β Blockers (%) | 167 (62) | 124 (62) | 41 (64) | 2 | 0.83 |
| Smoking (%) | 44 (16) | 28 (14) | 14 (22) | 2 | 0.06 |
| No AF | n = 1,403 | n = 1,110 | n = 276 | n = 17 | |
| Age (yrs) | 49.5 ± 14.7 | 47.6 ± 15 | 49 ± 15 | 42.4 ± 18 | 0.67 |
| Men (%) | 639 (46) | 520 (47) | 113 (41) | 6 (35) | 0.15 |
| Body mass index (kg/m 2 ) | 28.8 ± 7.8 | 28.7 ± 8.5 | 28.7 ± 8.5 | 23.5 ± 5.3 | 0.50 |
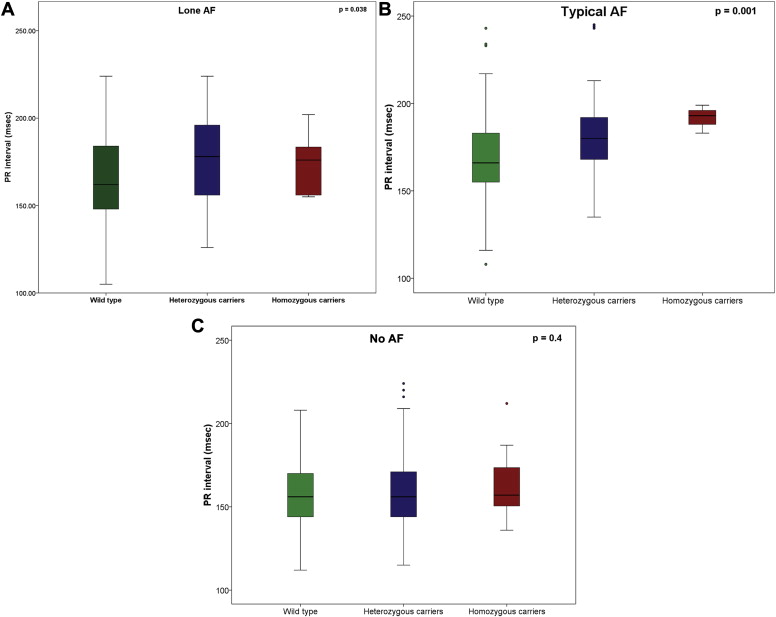
Median (interquartile range) PR intervals (in milliseconds) for lone AF, typical AF, any AF, and controls with no AF were 168 (152 to 188), 171 (156 to 187), 169 (155 to 188), and 156 (144 to 170), respectively. There were statistically significant differences in median PR intervals between lone AF and controls, typical AF and controls, any AF and controls (p <0.001 for each comparison), but not between lone and typical AF (p = 0.22). In univariate analysis, PR-interval duration differed by rs2200733 genotype in lone and typical AF ( Table 2 ). Among patients with lone AF, median (interquartile range) PR-interval durations were 162 (148 to 184), 178 (157 to 196), and 176 ms (156 to 181) for CC, CT, and TT, respectively (p = 0.038; Figure 1 ). In the typical AF group, median (interquartile range) PR-interval durations were 166 (155 to 183), 180 (168 to 192), and 196 ms (188 to 206) for CC, CT, and TT, respectively (p = 0.001; Figure 1 ). In univariate analysis, PR intervals did not differ by genotype in controls (median [interquartile range] 156 [144 to 169], 156 [144 to 170], and 170 ms [149 to 176] for CC, CT, and TT, respectively, p = 0.4; Figure 1 ).
| Cohort | Total | CC | CT | TT | p |
|---|---|---|---|---|---|
| Lone AF | 168 (152–188) | 162 (148–184) | 178 (157–196) | 176 (156–181) | 0.038 |
| Typical AF | 171 (156–187) | 166 (155–183) | 180 (168–192) | 196 (188–206) | 0.001 |
| No AF | 156 (144–170) | 156 (144–169) | 156 (144–170) | 170 (149–176) | 0.4 |