Fig. 5.1
Schematic representation of the transcriptional/translational feedback loop of the circadian core clock pathway in mammals. The central proteins BMAL1 and CLOCK form a heterodimer and bind E-box elements in the promoter of period (Per1/2) and cryptochrome (Cry1/Cry2) genes. PER and CRY can be degraded by the 26S proteasome in the cytoplasm after ubiquitination (U) by β-TRCP1/2 after casein kinase ε mediated phosphorylation (P). If not degraded in the cytoplasm, PER and CRY can dimerize and shuttle to the nucleus, where they inhibit their own transcription via blocking BMAL1:CLOCK transcriptional activity. After gradual phosphorylation, the PER/CRY complex is ubiquitinated by the F-Box protein FBXL3, and degraded in the nucleus. This lifts the repression on BMAL1:CLOCK, resulting in a new transcriptional cycle. In a secondary feedback loop, the BMAL1:CLOCK dimer drives transcription of Rev-erbα/β and RORα/β. Their proteins shuttle to the nucleus where they inhibit and activate Bmal1 transcription, respectively via competing for a Rev-erb response element (RRE) in the Bmal1 promoter. The result of one cycle, which takes approximately 24 h is the rhythmic transcriptional activation of clock-controlled genes (CCGs) by BMAL1:CLOCK. An oscillatory SIRT1 activity cycle integrates metabolism into the circadian clock. For additional information see text. Stars: histone tails
Transcription of two groups of CCGs, named Period (Per) genes and Cryptochrome (Cry) genes , are activated by BMAL1 and CLOCK. These output genes constitute the negative branch of the first autoregulatory feedback loop. Their necessary role in the clock pathway is underscored by the fact that Cry1/Cry2 double knockout mice also show a complete loss of rhythmicity (van Der Horst et al. 1999; Vitaterna et al. 1999). PER and CRY proteins accumulate and dimerize in the cytoplasm, where their presence is tightly regulated. They can either be stabilized by phosphatase1 (PPI1) (Gallego et al. 2006) or phosphorylated by casein kinases (CKε/δ) (Keesler et al. 2000; Lowrey et al. 2000; Camacho et al. 2001) resulting in active degradation. For CRY1 this process occurs via ubiquitination by F-box and leucine-rich repeat protein 21 (FBXL21) (Dardente et al. 2008; Hirano et al. 2013; Yoo et al. 2013). Phosphorylated PER2 can be polyubiquitinated by β-TRCP1/2 resulting in proteasomal degradation (Eide et al. 2005). In general, dimerization of PER:CRY in the cytoplasm protects both proteins from degradation (Yagita et al. 2002). Upon stabilization, the PER:CRY dimer translocates into the nucleus (Yagita et al. 2000) forming a nuclear complex (Brown et al. 2005). There, the dimer binds to the NuRD (nucleosome remodeling deacetylases) transcriptional repressor complex and directs NuRD to BMAL1:CLOCK (Kim et al. 2014). Then, a fully functional NuRD repressing complex is established, resulting in the inhibition of BMAL1:CLOCK driven transcription. Through this, PER and CRY inhibit multiple CCGs as well as their own transcription (Gekakis et al. 1998; Kume et al. 1999; Griffin et al. 1999). PER initiates this negative feedback loop by functioning as a molecular scaffold, that brings CRY into contact with BMAL1:CLOCK (Chen et al. 2009). In the nucleus CRY1 and CRY2 can be degraded by the proteasome, facilitated by F-box type E3 ubiquitin ligase (FBXL3 ). Its function is nicely illustrated by impaired degradation of CRY, as seen in the overtime (Ovtm) mutant, that causes enhanced inhibition of BMAL1:CLOCK-based transcription, resulting in period lengthening (Siepka et al. 2007). Upon decrease of nuclear PER and CRY levels, BMAL1:CLOCK inhibition is released, and a new transcriptional activation cycle of CCGs can start.
An additional layer of transcriptional control of the circadian clock comprises the orphan nuclear receptors ROR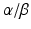 and REV-ERB
and REV-ERB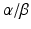 , encoded by Ror
, encoded by Ror 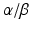 and Nr1d1/2 respectively. The BMAL1:CLOCK dimer mediates their transcription via binding to their E-box, initiating a second feedback loop (Preitner et al. 2002; Sato et al. 2004; Guillaumond et al. 2005). Both RORs and REV-ERBs compete for retinoic acid-related orphan receptor response element (RORE ) binding sites , also termed RRE (RevErbA response element), within the Bmal1 promoter (Harding and Lazar 1993; Ueda et al. 2002). Binding of RORs drives transcription of Bmal1 while REV-ERBα inhibits its transcription (Preitner et al. 2002; Akashi and Takumi 2005; Guillaumond et al. 2005). Although Rorα mRNA levels are only slightly oscillating, RORα is necessary for rhythmic Bmal1 expression (Akashi and Takumi 2005).
and Nr1d1/2 respectively. The BMAL1:CLOCK dimer mediates their transcription via binding to their E-box, initiating a second feedback loop (Preitner et al. 2002; Sato et al. 2004; Guillaumond et al. 2005). Both RORs and REV-ERBs compete for retinoic acid-related orphan receptor response element (RORE ) binding sites , also termed RRE (RevErbA response element), within the Bmal1 promoter (Harding and Lazar 1993; Ueda et al. 2002). Binding of RORs drives transcription of Bmal1 while REV-ERBα inhibits its transcription (Preitner et al. 2002; Akashi and Takumi 2005; Guillaumond et al. 2005). Although Rorα mRNA levels are only slightly oscillating, RORα is necessary for rhythmic Bmal1 expression (Akashi and Takumi 2005).
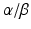 and REV-ERB
and REV-ERB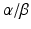 , encoded by Ror
, encoded by Ror 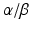 and Nr1d1/2 respectively. The BMAL1:CLOCK dimer mediates their transcription via binding to their E-box, initiating a second feedback loop (Preitner et al. 2002; Sato et al. 2004; Guillaumond et al. 2005). Both RORs and REV-ERBs compete for retinoic acid-related orphan receptor response element (RORE ) binding sites , also termed RRE (RevErbA response element), within the Bmal1 promoter (Harding and Lazar 1993; Ueda et al. 2002). Binding of RORs drives transcription of Bmal1 while REV-ERBα inhibits its transcription (Preitner et al. 2002; Akashi and Takumi 2005; Guillaumond et al. 2005). Although Rorα mRNA levels are only slightly oscillating, RORα is necessary for rhythmic Bmal1 expression (Akashi and Takumi 2005).
and Nr1d1/2 respectively. The BMAL1:CLOCK dimer mediates their transcription via binding to their E-box, initiating a second feedback loop (Preitner et al. 2002; Sato et al. 2004; Guillaumond et al. 2005). Both RORs and REV-ERBs compete for retinoic acid-related orphan receptor response element (RORE ) binding sites , also termed RRE (RevErbA response element), within the Bmal1 promoter (Harding and Lazar 1993; Ueda et al. 2002). Binding of RORs drives transcription of Bmal1 while REV-ERBα inhibits its transcription (Preitner et al. 2002; Akashi and Takumi 2005; Guillaumond et al. 2005). Although Rorα mRNA levels are only slightly oscillating, RORα is necessary for rhythmic Bmal1 expression (Akashi and Takumi 2005).A last transcriptional loop involved in circadian clock oscillation consists of a number of proline and acidic amino acid-rich basic leucine zipper (PAR bZIP ) transcription factors regulating circadian gene expression via binding D-box (DBP response element) elements (TTATG(C/T)AA) (Falvey et al. 1996). Albumin D-site-binding protein (DBP), thyrotroph embryonic factor (TEF ), and hepatic leukemia factor (HLF ) contribute to positive regulation, whereas nuclear factor interleukin 3 regulated (NFIL3 or E4BP4) provides negative regulation. Although this accessory loop is not strictly necessary for circadian oscillations, it provides robustness and precision to the period. An overview of the described core clock pathway is depicted in Fig. 5.1.
5.2.2 Epigenetic Regulation of the Circadian Clock
Besides the complex transcriptional/translational control system, circadian rhythmicity is also regulated by epigenetic mechanisms . Epigenetic control includes methylation of the DNA at CpG islands, non-coding RNAs and posttranslational modifications of histones. All of these epigenetic mechanisms have been implicated in driving and fine-tuning circadian rhythmicity in gene expression.
CLOCK , one of the core circadian rhythm proteins, can function as a histone acetyl transferase (HAT) that acetylates histone H3 on its lysine 9 (H3K9) and lysine 14 (H3K14) amino acid residues (Doi et al. 2006). H3K9ac and H3K14ac are both markers for permissive transcription. Therefore, BMAL1:CLOCK also regulates transcription of CCGs via modifying their histones (Etchegaray et al. 2003). This function is being neutralized by several histone deacetylases (HDACs), as described in more detail by Steven Brown (2011).
In search for these balancing HDACs , Sirtuin 1 (SIRT1) was discovered by the group of Sassone-Corsi as a protein that counteracts the HAT function of CLOCK. In general, Sirt1 is well studied in the context of aging, resistance to cellular stress, metabolism, inflammation and proliferation (Bordone and Guarente 2005). Additionally, SIRT1 is now known to deacetylate the proteins BMAL1, PER2 (Asher et al. 2008) and histone 3 (H3) on the promoter of clock output genes like Dbp (Nakahata et al. 2008). The deacetylating activity of SIRT1 is NAD+ (nicotinamide adenine dinucleotide) dependent and circadian. In the absence of de novo NAD+ biosynthesis , NAD+ needs to be replenished to avoid cell death. This goes via the NAD+ salvage pathway, where the by-product of NAD+ usage, NAM (nicotinamide) is reconverted into usable NAD+ via NMN (nicotinamide mononucleotide). In this process, NAMPT (NAM phosphoribosyltransferase) is the rate-limiting enzyme. As the expression of NAMPT itself is under circadian control, NAD+ also oscillates as an available metabolite. By this, SIRT1 links the metabolic state of a cell with the epigenetic control of the clock gene transcription pathway. As a rheostat of the circadian clock, SIRT1 mainly controls the amplitude of CCGs (Sassone-Corsi 2012).
Besides acetylation, methylation of histones is important to mediate circadian rhythmicity in gene expression. Histone H3 lysine trimethylation (H3K4me3) is a mark consistently associated with circadian transitions of the chromatin fiber, controlling CCG expression. One of these histone methyl transferases (HMTs) is mixed lineage leukemia 1 (MLL1) that can recruit the BMAL1:CLOCK dimer to the DNA of target genes and cause rhythmic H3K4 trimethylation (Katada and Sassone-Corsi 2010). Histone H3 lysine 27 trimethylation (H3K27me3) on the other hand is a repressive epigenetic mark and has been shown to play a counteracting role in circadian regulation of gene expression. The Per1 promoter for example shows rhythmic H3K27me3 marks, mediated by EZH2. The counteracting enzymes, histone demethylases , also play a role in circadian rhythms, where JARID1A is a known demethylase that inhibits HDAC1 and boosts BMAL1:CLOCK facilitated transcription of Per genes. Dynamic interaction between HDAC1 and JARID1A correlates with proper histone acetylation at the Per promoters (DiTacchio et al. 2011). The same holds true for LSD1, another histone demethylase whose activity depends on circadian phosphorylation by PKCα (Nam et al. 2014).
5.2.3 Additional Regulatory Systems Fine-Tune Circadian Rhythmicity
As described above, the genetic and epigenetic mechanisms underlying the circadian clock are quite complex and have been studied extensively. In addition to all this, a handful of papers describe even more ways of circadian regulation of the core clock pathway. Most of these additional mechanisms enhance robustness of the clock, rather than being truly essential for rhythmicity. A first example of such an extra clock dimension is the oscillating cold inducible RNA-binding protein (CIRP ). CIRP is a RNA binding protein, and is regulated by circadian rhythmicity in temperature oscillations. CIRP binds to Clock mRNA and stabilizes it, linking temperature to the circadian clock (Morf et al. 2012). Next, cyclic alternative splicing (McGlincy et al. 2012) as well as light inducible alternative splicing (Preußner et al. 2014) are common factors that regulate circadian rhythms. Third, rhythmic polyadenylation that stabilizes mRNA molecules, facilitates circadian rhythmicity in protein translation (Kojima et al. 2012). Furthermore, fluctuations in m(6)A-RNA methylation also affect the circadian transcriptome (Fustin et al. 2013). Finally, non-coding RNAs such as anti-sense RNAs and miRNAs affect circadian rhythms. This is demonstrated by a disrupted circadian transcriptome in cells that lack Dicer, a gene involved in miRNA processing (Chen et al. 2013; Du et al. 2014). In conclusion, the circadian clock is achieved by a complex orchestrated interplay between genetics, epigenetics and translational processes.
5.3 Circadian Rhythms in Tissue/Organ Physiology
5.3.1 Tissue Specific Control of Clock Output
The core clock machinery, as studied extensively in the SCN, is conserved in all peripheral organs. The basic signaling between the core oscillator and peripheral clocks involves both neuronal and humoral signals, such as melatonin. However, peripheral clocks can also respond independently to environmental cues such as body temperature and food metabolites (Damiola et al. 2000; Stokkan et al. 2001) (Fig. 5.2). Nonetheless, there are significant differences between each tissue in the relative contributions of the clock components, as well as in the output pathways that are under their control. These endogenous cellular clocks drive extensive rhythms of gene transcription, with 3–10 % of all mRNAs in a given tissue showing diurnal rhythms (Akhtar et al. 2002; Duffield et al. 2002; Miller et al. 2007). However, the genes that are under circadian control are largely non-overlapping in different tissues. This tissue specificity reflects the need for temporal control of the cellular physiology relevant to each unique cell type. As a result, the circadian clock exerts extensive control over many unique biological processes .
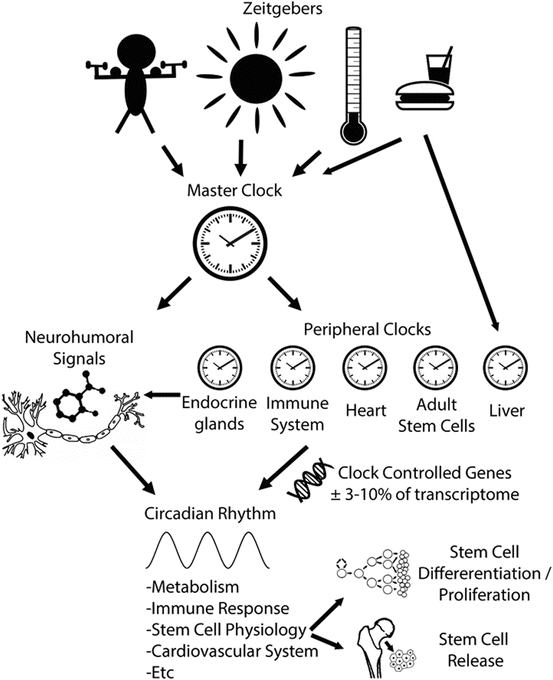

Fig. 5.2
The circadian clock regulates rhythmic body physiology. A master clock in the brain and peripheral clocks in almost every cell/tissue of the body drive circadian rhythmicity. Clocks are entrained by different factors/zeitgebers such as light and food. These rhythms are propagated by electrical and neurohumoral signals resulting in a 24-h rhythmic expression of clock-controlled genes. These genes render circadian rhythmicity in many functional processes ensuring proper body physiology and regeneration
5.3.2 Transgenic Animals Provide Novel Insights to Better Understand the Clock
Tissue-specific transgenic mice have been used to address the precise functions of peripheral clocks in physiological processes. For example, in liver-specific Bmal1 KO mice there is a loss of rhythmicity of glucose regulatory genes, which leads to an accelerated glucose clearance during the course of the daily feeding cycle (Lamia et al. 2008). In the adrenal glands, many genes involved in the biosynthesis of corticosterone are clock-controlled. Therefore the tissue specific disruption of Bmal1 interrupts the ability of the organ to maintain proper oscillatory secretion of corticosterone (Son et al. 2008). In pancreatic islets, the circadian clock helps regulating glucose-stimulated insulin secretion, the loss of which impairs glucose tolerance because of β-cell dysfunction (Marcheva et al. 2010).
The peripheral clock also plays a profound role in the cardiovascular system (Durgan and Young 2010; Paschos and FitzGerald 2010). In blood vessels, regulation of vascular function and tone has shown to be regulated by circadian rhythms. For example, deletion of Bmal1 specifically in vascular endothelium leads to a reduction of blood pressure during the active phase of the day and increased heart rate throughout the 24-h cycle (Westgate et al. 2008). The ability of endothelial cells from Per2 mutant mice to proliferate and form vascular networks is substantially reduced, which is marked by increased senescence of the cells (Wang et al. 2008). In vivo, Per2 mutant mice show decreased angiogenesis, as blood flow is impaired and combined with reduced recovery in response to ischemia characterized by a smaller increase in vessel formation. Lastly, Westgate et al. (2008) showed that the time to thrombotic vascular occlusion in response to a photochemical injury displays diurnal variation. Platelet aggregation factors, plasminogen activator inhibitor and tissue plasminogen activator, produced by the vascular endothelium show diurnal variability throughout the day/night cycle. In this regard, the deletion of Bmal1 specifically in the vascular endothelium results in loss of the temporal pattern in susceptibility to thrombotic vascular occlusion .
5.3.3 Circadian Rhythms in the Heart
In the heart, rhythmic physiology has mostly been studied in mice harboring a dominant negative version of the CLOCK protein in cardiomyocytes specifically (CCM mice). In vivo radiotelemetry studies performed in wildtype (WT) and CCM mice for continuous 24-h monitoring of physical activity, revealed a reduction in heart rate in the CCM mice (despite identical physical activity), which was especially pronounced during the awake/active phase. In this regard, the circadian clock seems to influence the generation and propagation of electric signals between adjacent cells in the heart. Although the precise mechanism remains unknown for this phenotype, the expression of various ion channels was found to be clock dependent. Furthermore, Connexin 40, a gap junction protein critical in atrial-ventricular conduction, shows oscillatory expression in WT hearts, that is absent in CCM hearts (Bray et al. 2008).
The metabolism of the heart is crucial for its contractile function, which has to meet the daily demand for increased workload during the active phase of the day. The two major components fueling the contraction of the myocardium are fatty acids and glucose (Taegtmeyer 2000). Diurnal gene expression studies in CCM mice revealed that a large number of genes influencing triglyceride (fatty acids) and glycogen (glucose) metabolism are controlled by the clock (Bray et al. 2008). The circadian regulation of lipolysis is exhibited in the diurnal variation of total triglyceride levels and synthesis in mouse hearts that peaks near the end of the active phase. These rhythms are essentially absent in CCM hearts. Furthermore, CCM mice display an altered response to high fat diet, showing a role of the cardiomyocyte circadian clock in the regulation of nonoxidative fatty acid metabolism (Tsai et al. 2010). Similarly, epinephrine-induced glycogenolysis has a time of the day dependent activity in WT hearts. However, these diurnal rhythms in cardiac glycogen metabolism are suppressed in CCM hearts (Bray et al. 2008).
Furthermore, hearts from 22-month-old CCM mice exhibited increased ejection fraction, fractional shortening, and left ventricular mass compared to WT. These are all characteristic of physiological hypertrophy and a strong link that the intrinsic clock of the heart also regulates myocardial growth. Growth factors, such as insulin-like growth factor-1 (IGF-1), are at the basis of physiological hypertrophy and can signal through the PI3K-Akt pathway . In this regard, the phosphorylation status of key components of this signaling pathway (Akt, GSK3β, and p70S6K) all oscillate in hearts over the course of the day, and are continually elevated in CCM hearts (Durgan et al. 2010). Myocardial growth is also closely associated with protein synthesis, which involves initiation factors (eIFs), several of which have been found to be under cardiomyocyte circadian regulation (Bray et al. 2008).
In summary, the peripheral clocks allow for individualized rhythmic gene expression in order for organs to be able to anticipate their diurnal tasks. In the cardiac context, heart rate, metabolism, and growth are all parameters that are under tissue specific circadian control. This enables the heart to be used most efficiently during periods of activity and rest.
5.4 Ontology of Circadian Rhythms and the Role of Clocks in Cell Cycle
5.4.1 Circadian Clocks During Embryonic Development
While the embryo develops, conditions within the uterus vary throughout the day. Concentrations of glucose and other metabolites for example, are relatively high during daytime compared to at night. A mature circadian clock to anticipate to these diurnal differences is not yet present in the embryo, but develops during gestation.
The proteins of the circadian clock, including CLOCK, BMAL, PER, and CRY are already present in the unfertilized oocyte (Johnson et al. 2002). However, their expression is non-rhythmic and low compared to adult cells. During development in utero, the expression of those genes gradually increases until birth (Saxena et al. 2007). Around the period of mid- to end gestation, diurnal oscillations of the core clock proteins commence. The amplitude of expression increases until the end of pregnancy and upon delivery, there is a 12-h phase shift, reversing 24-h clock gene expression (Saxena et al. 2007). In some species, such as rodents, development of the molecular clock even continues after birth.
Circadian clocks rely on Zeitgebers for synchronization to external surroundings. Light for example, is received by the retina and via internal cues, synchronizes circadian clocks within the body. The fetus does not directly receive light input. Also, many of the maternal Zeitgeber signals do not reach the fetus because they do not pass the placenta. Therefore, the regulation of molecular clocks within the fetus rely mainly on melatonin, which is produced by the maternal pineal gland and is able to pass the placenta (Reppert and Weaver 2002). The obtained 24-h rhythms play an important role in the physiology of the developing fetus. Metabolic activity and parameters such as breathing movements and heart rate show diurnal oscillations within the uterus (Visser et al. 1982; de Vries et al. 1987). When those rhythms are disrupted during gestation, this can have detrimental effects as indicated by shift work during pregnancy. This results in relatively small babies and animal experiments show that it may also lead to glucose intolerance and insulin resistance in newborns (Varcoe et al. 2011; Bonzini et al. 2011). A detailed overview of in utero development of circadian rhythms was previousl y published (Du Pré et al. 2014).
5.4.2 Oscillation of Core Clock Genes upon Differentiation of Embryonic Stem Cells
Previously, researchers have reported the absence of a clear functional core clock in pluripotent embryonic stem cells (ESCs ). Via bioluminescent reporter systems and the analysis of expression levels of clock genes, no clear circadian rhythms could be identified. However, when pushing these stem cells to leave their pluripotent, proliferative state through the withdrawal of leukemia inhibitory factor (LIF), circadian rhythms slowly started to emerge. By day 28, clear and robust rhythms were observed in D28 differentiated embryoid bodies (EBs) (Yagita et al. 2010; Umemura et al. 2013). The same holds true when stem cells are differentiated in a more directed way towards the neuronal lineage by addition of retinoic acid (Yagita et al. 2010; Kowalska et al. 2010). When differentiated cells are reprogrammed towards induced pluripotent stem cells (iPSCs), they lose their established oscillatory clock gene expression pattern. However, upon re-differentiation, they acquire an active circadian clock again. This suggests that an inherent circadian clock is linked to the differentiation status of a cell, and that this clock might be established during early phases of development (Brown 2014).
Albeit not clearly oscillating, most clock genes are expressed in ESCs. However, expression levels are different in comparison to differentiated cells. Per1, Per2 and Clock are expressed at a lower level, Cry2 is expressed at a higher level, and Bmal1 shows similar transcript levels (Umemura et al. 2014). Apart from being molecular oscillators, this implies a different function of clock genes in ESCs. A possible explanation for the lack of clock gene oscillation in ESCs, is proposed in a recent paper of the Yagita lab. They show that the decline of Importin α2, encoded by Kpn2α, plays a key role in the acquirement of a circadian clock in vitro. Importin α2 is a nuclear transporter and shuttles specific pluripotency factors, like OCT3/4, into the nucleus. Additionally, it keeps differentiation linked factors, like OCT6, in the cytoplasm. Through this, a pluripotent state is retained (Yasuhara et al. 2013). Importin α2 also keeps clock factors like PER1 and PER2 in the cytoplasm (Umemura et al. 2014). Therefore, the absence of a functional clock in murine ESCs might be accountable to the fact that PER2 stays in the cytoplasm in pluripotent cells. The start of circadian oscillation is proposed as the timed entry of PER2 into the nucleus (Yagita et al. 2000).
5.4.3 Role of the Circadian Clock in Cell Cycle
Not only the circadian clock system, but also the cell cycle is an important biological oscillator . Researchers have intensively studied whether both processes are related and what the possible link between them might be. The cell cycle is represented by four different consecutive phases that ultimately lead to the division of proliferative cells. The S-phase is the replicative phase, in which the DNA is being duplicated. Next, G2 is a growth phase in which the cell contains a fully doubled genome. This leads to the M- (or mitotic) phase, where the DNA is propagated to the newly formed cell through actual division. M-phase leads to G1, which is another growth phase after which the cell re-enters the S-phase. However, non-dividing (somatic) cells can escape the cell cycle at G1 and reside in G0. However, they can switch back to a proliferative state via re-entering the cell cycle. The transition between cell cycle phases is facilitated by cyclin dependent kinases (CDKs) and cyclins, establishing so called “cell cycle checkpoints ”.
In the group of F. Naef, the influence of the cell cycle on the circadian clock and vice versa, have been studied at a single cell level in NIH3T3 cells. In the absence of a synchronizing stimulus, cell division precedes the peak of Reverbα expression with 5 h, concluding a clear 1:1 coupling between the cell cycle and the circadian clock (Bieler et al. 2014). This process is termed “Phase locking ”. When disturbing the cell cycle length, the circadian period is impacted but the 1:1 coupling remains. In reverse, lengthening the circadian period is not affecting the cell cycle, concluding a clear unidirectional coupling between cell cycle and the circadian clock (Bieler et al. 2014). On the other hand, cells in organs are under influence of synchronizing stimuli (e.g. light, temperature, hormones, feeding). Therefore, Feillet et al. found that synchronizing in vitro cultured cells leads to the occurrence of two subpopulations in culture: one in which the 1:1 phase locking is sustained, and another in which two circadian clock periods coincide with three complete cell cycles (Feillet et al. 2014). This suggests that the cell cycle is synchronized via physiological stimuli with the circadian clock (Feillet et al. 2015). Both studies indicate a clear interaction between cell cycle and the circadian clock, with the dominating factor being dependent on the environment (Feillet et al. 2015).
At a molecular level, the circadian clock has been linked to the cell cycle in several ways. The two main cell cycle phase windows controlled by clock genes can be found at the G2/M and the G1/S phase transition. Cell cycle regulators such as Wee1 in the murine liver (Matsuo et al. 2003), p20 and p21 in a developing zebrafish embryo (Laranjeiro et al. 2013), c-Myc and cyclin D1 (Fu et al. 2002) are under control of several clock proteins and show circadian rhythmicity in their expression.
It has been shown that the key component of the negative limb of the clock pathway, PER2, can regulate the cell cycle via inhibiting c-MYC activity. C-MYC directly blocks cyclin D1 activity, which is a roadblock for G1/S phase transition in the cell cycle. Overexpression of Per2 concomitantly leads to cell cycle arrest in certain cancer cell lines and pushes these cells into apoptosis (Hua et al. 2006; Oda et al. 2009; Sun et al. 2010). Recently, a relatively new clock player termed NONO has also been implicated in cell cycle regulation. This multifunctional nuclear protein partners with PER2 and regulates expression of the cell cycle checkpoint gene p16-Ink4A in a circadian fashion. Since p16-INK4A facilitates G1 exit, NONO couples cell cycle to the circadian clock, which is argued to be a way to segregate cell proliferation from tissue organization in a time-based manner (Kowalska et al. 2013).
5.5 Circadian Rhythms in Adult Stem Cells
5.5.1 Regulation of Adult Stem Cells by Oscillatory Systems
Adult stem cells are multipotent stem cells that are present in adult organisms. They can proliferate and differentiate, but their potential is limited to specific cell types. Hematopoietic stem cells for example can differentiate into multiple blood cells, but not into skin cells. Adult stem cells are present in many organs. In some tissues they supply a constant renewal of cells (intestines, skin, blood), whereas in others they only become active after injury (heart, skeletal muscle). In the heart, adult stem cells of cardiac and non-cardiac origin are being investigated for their use in regenerative therapy.
24-hour rhythmicity plays an important role in adult stem cells. First, there are 24-h oscillations in stem cell mobilization and trafficking (Fig. 5.2). Hematopoietic stem cells (HSCs) in the circulation for example, show 24-h oscillations in both humans and mice (Laerum 1995; Méndez-Ferrer et al. 2008). In humans, there is a HSC release peak 5 h after the day-to-night transition. This rhythm is orchestrated by the circadian central clock, via diurnal noradrenalin secretion and local, sympathetic nerves to the bone marrow (Méndez-Ferrer et al. 2008). Oscillations in the number of circulating HSCs lead to 24-h oscillations in hematopoietic growth factors and the number of blood cells in the circulation (Laerum 1995; Gimble et al. 2009). As a consequence, 24-h oscillations are present in processes mediated by blood cells: diurnal rhythms in leukocytes for example cause oscillations in immune responses and rhythms in thrombocytes causes rhythms in coagulation time. Almost all adults have 24-h oscillations in HSC release. In aging and some diseases, however, these oscillations are dampened (Sletvold and Laerum 1988). Other adult stem cells in the circulation, such as endothelial progenitor cells , also show 24-h rhythms (Thomas et al. 2008).
5.5.2 Circadian Clock Gene Expression in Adult Stem Cells
Adult stem cells themselves also have molecular circadian clocks. Mesenchymal stem cells (MSCs), adult stem cells that can differentiate into cells of the mesoderm, such as osteoblasts, chondrocytes, and adipocytes, provide a good example of this. When MSCs are cultured in vitro, 24-h oscillations in gene expression of core clock genes such as Bmal1, Cry1, Per2/3, Rev-erbα/β, and Dbp are found, indicating that the peripheral circadian clock is present in these cells, independent of the presence of the central clock (Wu et al. 2008). Differences in expression between MSCs of old and young animals are observed (Yu et al. 2011). These rhythms seem to play an important role in physiology and pathophysiology. RORα for example, a component of the circadian clock present in MSCs, influences MSC differentiation (Meyer et al. 2000). When RORα is disrupted, bone mass parameters and bone geometry are impaired. Other studies have demonstrated that the circadian clock regulates bone metabolism, one of the functions of MSCs. Mice with altered circadian rhythms display bone remodeling and more than 26 % of the bone transcriptome exhibits circadian rhythmicity (Fu et al. 2005; Zvonic et al. 2007). Other functions of MSCs, such as adipose tissue homeostasis , also showed to be under circadian control (Guo et al. 2012).
Circadian rhythms in adult stem cells are associated with proliferation and differentiation (Fig. 5.2). Proliferation and differentiation are main characteristics of adult stem cells and vital for regenerative medicine. When MSCs are cultured in vitro and circadian rhythms are disrupted by translocation of CRY1 and PER1 from the nucleus to cytoplasm using laser irradiation, this leads to a change in differentiation from adipogenesis to osteogenesis (Kushibiki and Awazu 2008). In addition, genes that regulate stem cell proliferation and differentiation often show circadian rhythms in gene expression. There are genes, such as GSK3 and Sirt1, which play an important role in both the circadian molecular clock and proliferation (Trowbridge et al. 2006; Yang et al. 2006). In addition, the phase of circadian rhythms seems to play a role in differentiation. In human epidermal stem cells the phase of circadian rhythms determines proliferation predisposition (Janich et al. 2013). When differentiation is induced by TGFβ or calcium, the core clock gene phase determines whether the epidermal stem cells respond to these cues. Based on circadian clock phase, stem cells can be divided into ‘dormant’ or ‘active’ stem cells (Janich et al. 2011). Disruption of clock genes such as Bmal1 or Per1/2 leads to accumulation or depletion of dormant stem cells.
In tissues that do not face a significant amount of proliferation during homeostatic conditions, such as the liver, heart, and skeletal muscle, circadian rhythms play a role in proliferation and differentiation after injury. After muscle injury for example, satellite cells from the basal lamina of muscle fibers become active to regenerate the muscle (Chatterjee et al. 2014). When the core clock gene Bmal1 is disrupted, muscle regeneration is impaired because satellite cells cannot expand. In the liver, the circadian clock regulates cell cycle genes, which in turn regulate mitosis (Matsuo et al. 2003).
In summary, 24-h rhythms are important in the physiology of adult stem cells. The master clock orchestrates oscillations of adult stem cell numbers in the circulation, whereas peripheral clocks within stem cells regulate 24-h stem cell physiology. Specifically, circadian rhythms play an important role in proliferation and differentiation, both in normal physiology and after injury. Therefore, it would be interesting to determine if the dormancy/activity of cardiac stem cells is also clock mediated .
5.6 The Role of Circadian Rhythms in Tissue Regeneration for Cell-Based Therapy
The previous paragraphs have illustrated the importance of circadian rhythms in humans and specifically in the context of embryonic- and adult stem cells and their derivatives. Although research in this research field is scarce, it is likely that circadian rhythms will influence cell-based therapy.
< div class='tao-gold-member'>
Only gold members can continue reading. Log In or Register to continue
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


