Arrhythmogenic right ventricular dysplasia/cardiomyopathy (ARVD/C) is frequently associated with desmosomal mutations. However, nondesmosomal mutations may be involved. The aim of this study was to assess the contribution of a phospholamban (PLN) gene mutation to ARVD/C diagnosis according to the revised 2010 task force criteria (TFC). In 142 Dutch patients (106 men, mean age 51 ± 13 years) with proven ARVD/C (fulfillment of 2010 TFC for diagnosis), 5 known desmosomal genes (PKP2, DSP, DSC2, DSG2, and JUP) and the nondesmosomal PLN gene were screened. After genetic analysis, phenotypic characteristics of desmosomal versus PLN mutation carriers were compared. In 59 of 142 patients with ARVD/C (42%), no desmosomal mutation was found. In 19 of 142 patients (13%), the PLN founder mutation c.40_42delAGA (p.Arg14del) was identified. PLN mutation carriers more often had low-voltage electrocardiograms (p = 0.004), inverted T waves in leads V 4 to V 6 (p <0.001), and additional structural (p = 0.007) or functional (p = 0.017) left ventricular impairment, whereas desmosomal mutation carriers had more solitary right ventricular abnormalities. The revised TFC included 21 of 142 patients with proven ARVD/C who did not meet the 1994 TFC, including 7 PLN mutation carriers. In conclusion, there is a substantial contribution of PLN mutation to ARVD/C diagnosis by the 2010 TFC. In 32% of patients (19 of 59) with genetically unexplained proven ARVD/C, this nondesmosomal mutation was found. PLN mutation carriers have ARVD/C characteristics, including important right ventricular involvement, and additionally more often low-voltage electrocardiograms, inverted T waves in the left precordial leads, and left ventricular involvement.
Arrhythmogenic right ventricular (RV) dysplasia/cardiomyopathy (ARVD/C), also known as arrhythmogenic cardiomyopathy, is a hereditary desmosomal disease with RV and possibly also left ventricular (LV) involvement. ARVD/C is diagnosed according to international consensus–based task force criteria (TFC) for family history, depolarization and repolarization abnormalities, ventricular arrhythmias, global and/or regional dysfunction and structural abnormalities of the right ventricle, and tissue characterization. The TFC were revised in 2010, creating a new set of criteria that improved the diagnostic yield. In the revised 2010 TFC, a major criterion was assigned to the identification of a pathogenic gene mutation associated with ARVD/C. Pathogenic desmosomal gene mutations have been identified in approximately 60% of Dutch index patients. In a minority of patients with ARVD/C, a nondesmosomal mutation predisposes to the phenotype. The c.40_42delAGA (p.Arg14del) mutation in the nondesmosomal phospholamban (PLN) gene was first described in 2 distinct families with heart failure and ventricular tachyarrhythmias. Recently in The Netherlands, we identified the c.40_42delAGA founder mutation in a large series of patients diagnosed with dilated cardiomyopathy or ARVD/C. The aims of this study were (1) to assess the contribution of the PLN mutation to ARVD/C phenotype and diagnosis according to the 2010 TFC and (2) to evaluate the phenotype of ARVD/C patients with desmosomal mutations and those with the nondesmosomal PLN mutation.
Methods
A total of 153 Dutch Caucasian ARVD/C index patients from 6 university centers (mean age 51 ± 14 years, 114 [75%] men) diagnosed according to the 2010 TFC or at autopsy were included. A subset of this cohort has been described in previous studies by Cox et al and van der Zwaag et al. An index patient was the first in the family diagnosed with ARVD/C in whom deoxyribonucleic acid (DNA) analysis was started. All patients consented to clinical and DNA evaluation.
The diagnostic process included a detailed clinical and family history, physical examination, 12-lead electrocardiography (while off drugs, in 100%), 48-hour ambulatory electrocardiographic monitoring (in 70%), exercise testing (in 69%), and 2-dimensional transthoracic echocardiography (in 89%). In addition, magnetic resonance imaging (in 67%) with delayed enhancement analysis (in 30%), electrophysiologic study (in 62%), RV and LV cine angiography (in 51%), and endomyocardial biopsy (in 47%) were performed when indicated.
Autopsy was performed in 5 patients. The cardiac tissue specimens were stained with hematoxylin and eosin and Masson’s trichrome stain. Findings from endomyocardial biopsies were not included in the study, because of small numbers of diagnostic biopsies and assessment according to either the 1994 or 2010 TFC.
Genomic DNA was extracted from peripheral blood as described before. Direct sequence analysis of all coding regions and intron-exon boundaries of the desmosomal genes plakophilin-2 (PKP2), desmoplakin (DSP), plakoglobin (JUP), desmoglein-2 (DSG2), desmocollin-2 (DSC2), and the PLN gene was performed. In addition, multiplex ligation-dependent probe amplification analysis was performed to identify large PKP2 deletions (SALSA MLPA kit P168 ARVC-PKP2; MRC Holland, Amsterdam, The Netherlands). Primer sequences and polymerase chain reaction conditions are available on request.
Mutations were considered proven pathogenic when they were nonsense, frame-shift, splice-site, or exon deletion mutations and not identified as polymorphisms. For missense variants, the predictive algorithms sorting tolerant from intolerant and polymorphism phenotyping–2 were used. Missense variants were considered most likely pathogenic when the 2 algorithms predicted a deleterious effect (sorting tolerant from intolerant: tolerance index score ≤0.05; polymorphism phenotyping–2: probably pathogenic classification) and variants were absent in 200 Dutch Caucasian control subjects. When available, cosegregation analysis data were additionally taken into account.
For phenotype comparison of desmosomal and PLN mutation carriers, only those patients with proven pathogenic or most likely pathogenic mutations were labeled as pathogenic mutation carriers. Patients carrying unclassified variants in the desmosomal genes were labeled together with those patients in whom no mutation or variant could be identified, as without pathogenic mutation. After clinical and genetic screening, patients were divided into 2 groups: pathogenic (1) desmosomal and (2) PLN mutation carriers. The occurrence of 2010 TFC and RV and LV involvement was assessed in the 2 mutation groups. RV involvement was considered positive in the presence of ≥1 of the following: negative T waves in leads V 1 to V 3 , RV wall motion abnormalities (akinesia or dyskinesia) and/or delayed enhancement in the right ventricle on imaging studies, and an RV ejection fraction ≤45%. LV involvement was considered positive in the presence of ≥1 of the following: negative T waves in leads V 4 to V 6 , LV wall motion abnormalities (akinesia or dyskinesia) and/or delayed enhancement in the left ventricle on imaging studies, and an LV ejection fraction <50%. Finally, the inclusion of the PLN mutation and associated phenotype in ARVD/C diagnosis by the revised TFC was measured.
Continuous variables were compared using Student’s t test. Categorical variables were analyzed using contingency tables and Fisher’s exact test. Descriptive statistics are reported as mean ± SD. A p value ≤0.05 was considered statistically significant. Results of phenotype comparison are shown as odds ratio estimates with associated confidence intervals. Correction for multiple testing was performed using the Holm-Bonferroni correction. All analyses were performed with PASW Statistics version 20.0 (SPSS, Inc., Chicago, Illinois) and R (R Foundation for Statistical Computing, Vienna, Austria).
Results
All 153 patients with proven (definite) ARVD/C underwent screening of desmosomal genes PKP2, DSP, JUP, DSG2, and DSC2. In 11 of 153 patients, additional screening of PLN was impossible because of insufficient DNA or lack of patient permission. Thus, 142 of 153 patients underwent screening of the desmosomal genes and the nondesmosomal PLN gene. Pathogenic desmosomal mutations were identified in 83 of 142 patients (58%; Table 1 ). Of 142 patients with proven AVRD/C, 19 (13%) carried the PLN c.40_42delAGA (p.Arg14del) mutation. No other mutations besides this founder mutation were identified in the PLN gene. More important, in all 19 PLN mutation carriers, this was the only pathogenic mutation found. Consequently, PLN c.40_42delAGA explained nearly 1/3 (19 of 59 [32%]) of the until then genetically unexplained cases of ARVD/C. The nondesmosomal PLN gene is at present, after the desmosomal PKP2 gene, the gene in which second most frequently a mutation is identified in Dutch patients with ARVD/C, and the c.40_42delAGA mutation is found more frequently than each individual desmosomal mutation.
| Gene (No. of Patients With Mutation) | Deoxyribonucleic Acid Change | Protein Change | Frequency |
|---|---|---|---|
| PKP2 (n = 74) | Deletion exons 1–4 | p.(?) | 1/74 |
| Deletion exons 1–14 | p.(?) | 1/74 | |
| Deletion exon 10 | p.(?) | 1/74 | |
| c.148_151del | p.(Thr50fs) | 1/74 | |
| c.235C>T | p.(Arg79 ∗ ) | 9/74 | |
| c.258T>G | p.(Tyr86 ∗ ) | 1/74 | |
| c.397C>T | p.(Gln133 ∗ ) | 6/74 | |
| c.917_918del | p.(Pro318fs) | 3/74 | |
| c.1211dup | p.(Val406fs) | 11/74 | |
| c.1369_1372del | p.(Gln457 ∗ ) | 3/74 | |
| c.1378G>A | p.(Val445fs) | 1/74 | |
| c.1848C>A | p.(Tyr616 ∗ ) | 4/74 | |
| c.1951C>T | p.(Arg651 ∗ ) | 1/74 | |
| c.2028G>A | p.(Trp676 ∗ ) | 1/74 | |
| c.2034G>A | p.(Trp678 ∗ ) | 1/74 | |
| c.2062T>C | p.(Ser688Pro) | 1/74 | |
| c.2146-1G>C | p.(Met716fs) | 6/74 | |
| c.2386T>C | p.(Cys796Arg) | 10/74 | |
| c.2421C>A | p.(Tyr807 ∗ ) | 1/74 | |
| c.2489+1G>A | p.(Lys768fs) | 5/74 | |
| c.2489+4A>C | p.(Lys768fs) | 4/74 | |
| c.2509del | p.(Ser837fs) | 1/74 | |
| c.2544G>A | p.(Trp848 ∗ ) | 1/74 | |
| DSP (n = 1) | c.1982A>T | p.(Asn661Ile) | 1/1 |
| JUP (n = 0) | |||
| DSG2 (n = 6) | c.137G>A | p.(Arg46Gln) | 2/6 |
| c.614C>T | p.(Pro205Leu) | 1/6 | |
| c.378+2T>G | p.(Ile73_Leu126del) | 1/6 | |
| c.874C>T | p.(Arg292Cys) | 1/6 | |
| c.1003A>G (homozygous) | p.(Thr335Ala) | 1/6 | |
| DSC2 (n = 3) | c.608G>A (homozygous) | p.(Arg203His) | 1/3 |
| c.942+3A>G | p.(?) | 1/3 | |
| c.943-1G>A | p.(?) | 1/3 |
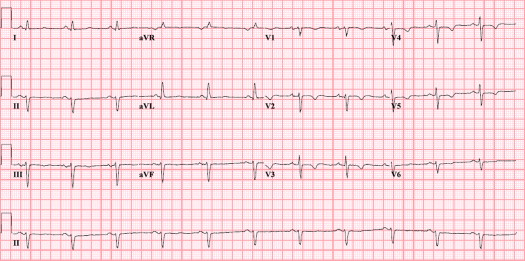
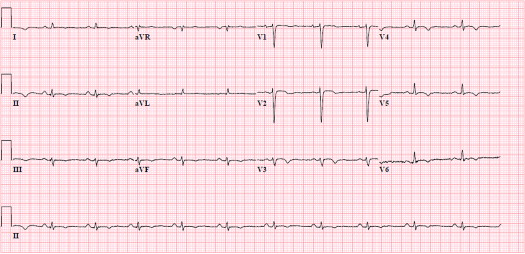
Baseline characteristics were similar in desmosomal and PLN mutation groups. Mean follow-up duration was 12 ± 9 years in the desmosomal mutation carriers and 10 ± 5 years in PLN mutation carriers (p = 0.258). Findings are listed in Table 2 . Sustained ventricular tachyarrhythmia with left bundle branch block morphology (i.e., originating from the right ventricle) and RV dysfunction were present in most patients with ARVD/C, regardless of the related desmosomal or PLN mutation. Desmosomal mutation carriers were more often characterized by negative T waves in right precordial leads V 1 to V 3 and beyond (a major criterion; Figure 1 ) in comparison with PLN mutation carriers. Nevertheless, this characteristic was also present in nearly half of PLN mutation carriers, as demonstrated by Figure 2 . PLN mutation carriers more often had low-voltage electrocardiograms and particularly negative T waves in left precordial leads V 4 to V 6 (a minor criterion; Figure 3 ). The minor criterion of >500 ventricular extrasystoles during 24-hour Holter monitoring was furthermore present in most patients with ARVD/C with the PLN mutation. Additional LV structural (wall motion abnormalities) and functional (reduced LV ejection fraction) abnormalities were considerably more often observed in PLN mutation carriers. Accordingly, the combined parameter of RV involvement was a common finding in the 2 groups ( Figure 4 ), whereas additional LV involvement was significantly more frequent a phenotypic expression of the PLN mutation ( Figure 4 ). Assessment of odds ratio estimates of unadjusted p values and associated confidence intervals seemed to confirm all aforementioned phenotypic differences between the 2 mutation groups, except for late potentials. In addition, even after statistical correction for multiple testing by the conservative Holm-Bonferroni correction, negative T waves in left precordial leads V 4 to V 6 and LV involvement stood out as significantly different between desmosomal and PLN mutation carriers.
| Clinical Parameter | Desmosomal Mutation (n = 83) | PLN Mutation (n = 19) | p Value | OR Estimate | Lower Limit | Upper Limit | Adjusted p Value ∗ |
|---|---|---|---|---|---|---|---|
| Men | 60 (72%) | 12 (63%) | 0.419 | 1.52 | 0.45 | 4.82 | 1.000 |
| Age (yrs) | 50 ± 14 | 52 ± 11 | 0.663 | −1.28 | −7.21 | 4.65 | 1.000 |
| Family history | |||||||
| Family history | 1/83 (1%) | 0/19 (0%) | 1.000 | Inf | 0.01 | Inf | 1.000 |
| Autopsy | 6/83 (7%) | 1/19 (5%) | 1.000 | 1.40 | 0.15 | 68.03 | 1.000 |
| Premature SCD | 5/83 (6%) | 4/19 (21%) | 0.060 | 0.25 | 0.05 | 1.38 | 0.950 |
| Electrocardiography | |||||||
| TAD ≥55 ms | 44/83 (53%) | 11/19 (58%) | 0.801 | 0.82 | 0.26 | 2.51 | 1.000 |
| Epsilon waves | 10/83 (12%) | 1/19 (5%) | 0.685 | 2.45 | 0.31 | 112.82 | 1.000 |
| Late potentials | 26/61 (43%) | 2/16 (13%) | 0.039 † | 5.11 | 1.03 | 50.19 | 0.660 |
| Low voltages | 16/73 (22%) | 11/19 (58%) | 0.004 † | 0.21 | 0.06 | 0.68 | 0.089 |
| Inverted T waves in leads V 1 –V 3 | 63/83 (76%) | 8/19 (42%) | 0.006 † | 4.26 | 1.35 | 14.11 | 0.127 |
| Inverted T waves in leads V 4 –V 6 | 6/83 (7%) | 10/19 (53%) | <0.001 † | 0.07 | 0.02 | 0.28 | <0.001 † |
| Arrhythmias | |||||||
| >500 VES/24 h | 30/55 (55%) | 14/16 (88%) | 0.020 † | 0.18 | 0.02 | 0.88 | 0.360 |
| LBBB VT | 75/83 (90%) | 15/19 (79%) | 0.229 | 2.47 | 0.48 | 10.78 | 1.000 |
| LBBB VT, superior axis | 49/83 (59%) | 8/19 (42%) | 0.207 | 1.97 | 0.64 | 6.29 | 1.000 |
| RBBB VT | 11/72 (15%) | 5/19 (26%) | 0.311 | 0.51 | 0.13 | 2.17 | 1.000 |
| Structural | |||||||
| Structural major ‡ | 60/83 (72%) | 14/19 (74%) | 1.000 | 0.93 | 0.24 | 3.15 | 1.000 |
| RV WMA | 65/73 (89%) | 15/19 (79%) | 0.262 | 2.15 | 0.42 | 9.39 | 1.000 |
| RV DE | 19/26 (73%) | 5/7 (71%) | 1.000 | 1.08 | 0.08 | 8.80 | 1.000 |
| LV WMA | 9/71 (13%) | 8/19 (42%) | 0.007 † | 0.20 | 0.06 | 0.75 | 0.144 |
| LV DE | 10/21 (48%) | 3/7 (43%) | 1.000 | 1.20 | 0.16 | 10.34 | 1.000 |
| Functional | |||||||
| RV EF ≤45% | 23/26 (89%) | 10/11 (91%) | 1.000 | 0.77 | 0.01 | 11.06 | 1.000 |
| LV EF <50% | 12/38 (32%) | 11/16 (69%) | 0.017 † | 0.22 | 0.05 | 0.86 | 0.320 |
| Ventricular involvement | |||||||
| RV involvement | 79/83 (96%) | 18/19 (95%) | 1.000 | 1.1 | 0.02 | 11.98 | 1.000 |
| LV involvement | 25/83 (30%) | 14/19 (74%) | 0.001 † | 0.16 | 0.04 | 0.52 | 0.026 † |
∗ Adjusted p value: corrected p value by Holm-Bonferroni correction.
† Statistically significant (p ≤0.05).
‡ Structural RV abnormalities accounting for a major criterion.


Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


