For many patients with atrial fibrillation, ventricular rate control with atrioventricular (AV) nodal blockers is considered first-line therapy, although response to treatment is highly variable. Using an extreme phenotype of failure of rate control necessitating AV nodal ablation and pacemaker implantation, we conducted a genome-wide association study (GWAS) to identify genomic modulators of rate control therapy. Cases included 95 patients who failed rate control therapy. Controls (n = 190) achieved adequate rate control therapy with ≤2 AV nodal blockers using a conventional clinical definition. Genotyping was performed on the Illumina 610-Quad platform, and results were imputed to the 1000 Genomes reference haplotypes. A total of 554,041 single-nucleotide polymorphisms (SNPs) met criteria for minor allele frequency (>0.01), call rate (>95%), and quality control, and 6,055,224 SNPs were available after imputation. No SNP reached the canonical threshold for significance for GWAS of p <5 × 10 −8 . Sixty-three SNPs with p <10 −5 at 6 genomic loci were genotyped in a validation cohort of 130 cases and 157 controls. These included 6q24.3 (near SAMD5/SASH1 , p = 9.36 × 10 −8 ), 4q12 ( IGFBP7 , p = 1.75 × 10 −7 ), 6q22.33 (C6orf174, p = 4.86 × 10 −7 ), 3p21.31 ( CDCP1 , p = 1.18 × 10 −6 ), 12p12.1 ( SOX5 , p = 1.62 × 10 −6 ), and 7p11 ( LANCL2 , p = 6.51 × 10 −6 ). However, none of these were significant in the replication cohort or in a meta-analysis of both cohorts. In conclusion, we identified several potentially important genomic modulators of rate control therapy in atrial fibrillation, particularly SOX5 , which was previously associated with heart rate at rest and PR interval. However, these failed to reach genome-wide significance.
Atrial fibrillation (AF) is the most common sustained clinical arrhythmia, affecting an estimated 6 million patients in the United States. Given the increasing incidence of AF observed over the past 2 decades, its prevalence in the United States is projected to increase to 12 to 16 million by the year 2050. For many patients with AF, pharmacologic therapy with atrioventricular (AV) nodal blockers is a first-line therapeutic strategy to control the ventricular rate and prevent symptoms and systolic heart failure associated with rapid ventricular rates. This is generally accomplished with the administration of calcium channel blockers, β-adrenergic blockers, and/or digitalis, which slow conduction in the AV node. Rate control therapy has been shown to be equivalent at preventing clinical outcomes to a rhythm control strategy, in which antiarrhythmic drugs and electrical cardioversions are used to maintain sinus rhythm. However, a substantial minority (∼30%) of patients fail rate control therapy, either due to persistent symptoms or inability to control the ventricular rate despite therapeutic doses of multiple AV nodal blockers. Genetic differences might explain some of the interindividual variability in the response to rate control therapy. We recently demonstrated that a single-nucleotide polymorphism (SNP) in the β1-adrenergic receptor gene (ADRB1) is associated with response to rate control therapy. Recent genome-wide association studies (GWASs) have identified genomic predictors of heart rate at rest in sinus rhythm and PR interval. These findings suggest that response to rate control therapy may also be genotype dependent. The goal of this study was to conduct a GWAS comparing patients with AF who responded effectively and ineffectively with ventricular rate control therapy. Given that the efficacy of rate control therapy is highly variable and is a complex clinical phenotype with multiple potential confounders, we chose an extreme phenotype for our analysis: failure of multiple AV nodal blockers necessitating AV node ablation and pacemaker implantation.
Methods
The discovery cohort consisted of patients in the Vanderbilt AF Registry (VAFR). As previously described, patients with AF have been recruited from Vanderbilt arrhythmia and general cardiology clinics, the emergency department, and inpatient services since April 2000. The replication cohort consisted of patients selected from the Vanderbilt University Medical Center DNA Biobank (BioVU). This resource consists of de-identified medical records of Vanderbilt inpatients and outpatients and DNA extracted from blood that is left over after routine laboratory testing and scheduled to be discarded. The Vanderbilt University Institutional Review Board has approved VAFR and BioVU. All patients in VAFR gave written informed consent. Given their de-identified nature, patient records and biologic samples in BioVU fall under the designation of “nonhuman subjects” under Title 45, Code of Federal Regulations, Part 46, and individual informed consent is not obtained. BioVU includes a mechanism to exclude subjects known to be a part of prospective registries such as VAFR; thus, samples from patients in VAFR were not included in the BioVU replication set.
Cases included patients in whom rate control therapy failed necessitating AV node ablation and permanent pacemaker implantation. Controls were patients with AF who achieved criteria for successful rate control (ventricular rate ≤80 beats/min at rest and not higher than 110 beats/min during a 6-minute walk test), as outlined in the Atrial Fibrillation Follow-up Investigation of Rhythm Management (AFFIRM) study, with ≤2 AV nodal blockers. The discovery cohort included 285 patients with persistent AF. Of these, 95 failed rate control therapy necessitating AV node ablation and pacemaker implantation (cases) and 190 were successfully rate controlled with ≤2 AV nodal blockers (controls). The validation cohort consisted of 130 cases and 157 controls defined with the same criteria. Baseline clinical characteristics are presented in Table 1 .
| Discovery | Validation | |||||
|---|---|---|---|---|---|---|
| Cases (N = 95) | Controls (N = 190) | p Value ∗ | Cases (N = 130) | Controls (N = 157) | p Value ∗ | |
| Age (years) | 69.2 ± 9.8 | 65.8 ± 10.3 | 0.22 | 73.4 ± 11.6 | 56.8 ± 11.3 | <0.001 |
| Male | 60% | 78% | 0.001 | 53.1% | 67.5% | 0.001 |
| Hypertension | 64% | 69% | 0.4 | 68% | 66% | 0.45 |
| Diabetes | 31% | 19% | 0.04 | 32% | 18% | 0.01 |
| Coronary artery disease | 32% | 30% | 0.77 | 72% | 14% | <0.001 |
| Myocardial infarction | 30% | 14% | 0.008 | 13% | 5.5% | 0.03 |
| Heart failure | 30% | 22% | 0.15 | 81% | 9.2% | <0.001 |
∗ Chi-square test for discrete variables, Mann-Whitney U test for continuous variables.
Genomic DNA was extracted from whole blood using standard techniques. Genotyping in the discovery cohort was performed for 620,902 variants with the Illumina 610-Quad platform (San Diego, California). We performed quality control of these data by excluding samples with >5% missing genotypes, and we estimated probability of identity by descent to check for family relationships, and agreement with database gender using X chromosome heterozygosity rates in PLINK. We dropped 2 samples for missing data, no samples for relationships above the first cousin level, and no samples for gender discordance. For SNPs within the set of high-quality samples, we dropped 43,807 SNPs for missing >5% genotypes, 54,558 monomorphic and rare SNPs with minor allele frequencies <1%, and 1,601 SNPs for Hardy-Weinberg equilibrium test p values <1 × 10 −6 . We also removed 2,718 palindromic SNPs to facilitate genotype imputation and removed 15,553 nonautosomal SNPs. In the discovery cohort, 502,665 SNPs met the prespecified criteria for inclusion in the analysis.
We imputed genotypes from the 1000 Genomes cosmopolitan reference panel of haplotypes using IMPUTE. A total of 6,055,224 genotyped and imputed SNPs with minor allele frequencies ≥5% and information scores >0.4 were included in primary discovery analyses. Logistic regression models were fit for each SNP using SNPTEST, regressing the response to rate control outcome onto genotype, age, and gender. To evaluate quality control, we calculated the genomic control parameter for tests at genotyped and imputed autosomal SNPs and λ = 1.025, suggesting that there were no strong unaccounted for biases or confounders ( Supplementary Figure 1 ).
We ranked SNPs by p value and conducted secondary analyses adjusting for the index or the most significant SNP in each genomic region within 1 megabase. We then selected a set of haplotype tag SNPs for the CEU reference population to tag a 100-kb region centered on each index SNP with r 2 ≥0.8 and minor allele frequencies ≥0.05 and genotyped the index and tag SNPs in the replication samples (130 BioVU cases and 157 BioVU controls) using the Sequenom MassArray genotyping platform (Sequenom, Inc, San Diego, California). A total of 63 SNPs at 6 genomic regions were selected for replication experiments ( Table 2 and Figure 1 ).
| Region | SNP | Nearest Gene | Effect/Referent | Effect Allele Frequency | Information | OR | 95% CI | p-Value |
|---|---|---|---|---|---|---|---|---|
| 6q24 | rs1534773 | SAMD5/SASH1 | T/C | 0.82 | 0.99 | 0.28 | (0.18–0.45) | 9.36 × 10 −8 |
| 4q12 | rs4242007 | IGFBP7 | C/G | 0.05 | 0.90 | 7.11 | (3.41–14.84) | 1.75 × 10 −7 |
| 6q22 | rs1512450 | C6orf174 | G/C | 0.68 | 0.99 | 0.36 | (0.24–0.55) | 1.11 × 10 −6 |
| 3p21 | rs55891698 | CDCP1 | T/C | 0.05 | 0.80 | 6.2 | (2.98–13.21) | 1.18 × 10 −6 |
| 12p12 | rs7967643 | SOX5 | T/C | 0.82 | 0.99 | 0.31 | (0.19–0.50) | 1.62 × 10 −6 |
| 7p11 | rs815929 | LANCL2 | A/C | 0.89 | 0.89 | 0.27 | (0.15–0.47) | 6.51 × 10 −6 |
Quality control procedures in the replication cohort were conducted as in the discovery cohort. No samples failed quality control. Two SNPs failed missing data filters. We then imputed the candidate regions to the 1000 Genomes cosmopolitan reference haplotypes with IMPUTE and conducted logistic regression analysis in the replication samples using SNPTEST. We then conducted fixed-effects inverse variance weighted meta-analysis with METAL to obtain combined evidence of effects and significance from discovery and replication samples.
Results
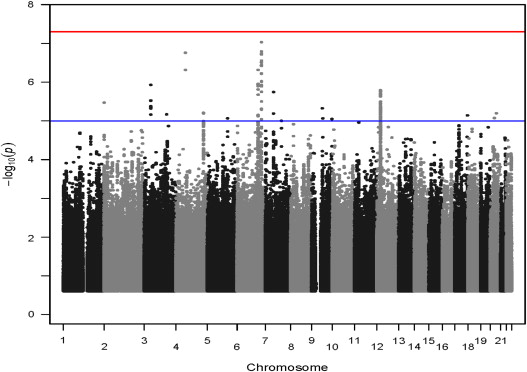
In the discovery analysis, SNPs in 6 genomic regions were associated with rate control therapy failure ( Table 2 ). No SNP reached the canonical threshold for significance for GWAS of p <5 × 10 −8 in the discovery analysis ( Figure 2 ). Multiple SNPs in the 6q24.3, 6q22.33, 4q12, 12p12.1, 3p21.31, and 7p11 regions were nominally significantly associated ( Figure 1 ).
None of the 63 SNPs genotyped in the replication cohort were significantly associated with failure of rate control therapy ( Table 3 ). Using the genotypes for these 63 SNPs, we imputed 2,792 SNPs. The region near the IGFBP7 gene on chromosome 4 was the only region with p values <0.05 in the replication cohort.
| SNP | Effect/Referent | Effect Allele Frequency | Information | OR | 95% CI | p-Value |
|---|---|---|---|---|---|---|
| rs4865181 | A/G | 0.22 | 0.96 | 0.57 | (0.38–0.85) | 0.006 |
| rs13119745 | T/C | 0.27 | 0.61 | 0.51 | (0.31–0.83) | 0.007 |
| chr4:57950946 | TA/T | 0.43 | 0.45 | 1.89 | (1.17–3.03) | 0.009 |
| rs6554428 | T/C | 0.55 | 0.90 | 1.55 | (1.11–2.15) | 0.010 |
| rs13108835 | C/A | 0.55 | 0.90 | 1.55 | (1.11–2.15) | 0.010 |
| rs11133483 | A/G | 0.57 | 0.97 | 1.54 | (1.11–2.13) | 0.010 |
| rs11133484 | G/A | 0.57 | 0.97 | 1.54 | (1.11–2.13) | 0.010 |
| rs7661524 | C/G | 0.55 | 0.91 | 1.54 | (1.11–2.14) | 0.010 |
| rs7681360 | A/G | 0.57 | 0.97 | 1.53 | (1.11–2.12) | 0.010 |
| rs11133482 | T/C | 0.57 | 0.97 | 1.53 | (1.11–2.12) | 0.010 |
| rs13129156 | G/A | 0.55 | 0.93 | 1.53 | (1.1–2.12) | 0.011 |
| rs9993153 | A/G | 0.55 | 0.92 | 1.53 | (1.1–2.13) | 0.011 |
| rs13147020 | G/A | 0.57 | 0.98 | 1.52 | (1.1–2.11) | 0.011 |
| rs7670481 | T/C | 0.54 | 0.83 | 1.55 | (1.1–2.19) | 0.013 |
| rs6825609 | A/G | 0.56 | 0.95 | 1.50 | (1.09–2.08) | 0.014 |
| rs4865182 | A/G | 0.24 | 1.00 | 0.62 | (0.43–0.91) | 0.015 |
| rs4075349 | T/C | 0.41 | 0.95 | 0.68 | (0.49–0.94) | 0.020 |
| rs1714007 | G/A | 0.65 | 1.00 | 1.49 | (1.06–2.08) | 0.020 |
| rs1714006 | G/A | 0.65 | 1.00 | 1.49 | (1.06–2.08) | 0.020 |
| rs1718829 | C/A | 0.65 | 0.99 | 1.49 | (1.06–2.08) | 0.020 |
A meta-analysis was performed for the association of the imputed SNPs with failure of rate control therapy in the discovery and replication cohort (225 cases and 347 controls). None of the SNPs were significantly associated with failure of rate control therapy, and the most significantly associated SNPs with p values <10 −4 were in the 6q24 region between the SAMD5 and SASH1 genes ( Table 4 ).
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


